Laura Eme
@lauraeme.bsky.social
2.2K followers
180 following
500 posts
Evolutionary biologist interested in the origin and early diversification of eukaryotes. Phylogenetics/(meta)genomics. Protists & Archaea. Reseach Director at Uni Paris-Saclay (France)/Associate Prof. Uni of Rhode Island (USA)
Posts
Media
Videos
Starter Packs
Reposted by Laura Eme
Laura Eme
@lauraeme.bsky.social
· Jun 26
Reposted by Laura Eme
Laura Eme
@lauraeme.bsky.social
· Jun 23
Laura Eme
@lauraeme.bsky.social
· Jun 22
Laura Eme
@lauraeme.bsky.social
· Jun 22
Laura Eme
@lauraeme.bsky.social
· Jun 22
Laura Eme
@lauraeme.bsky.social
· Jun 22
Laura Eme
@lauraeme.bsky.social
· Jun 19
Laura Eme
@lauraeme.bsky.social
· Jun 19
Laura Eme
@lauraeme.bsky.social
· Jun 19
Laura Eme
@lauraeme.bsky.social
· Jun 17
Laura Eme
@lauraeme.bsky.social
· Jun 17
Laura Eme
@lauraeme.bsky.social
· Jun 17
Laura Eme
@lauraeme.bsky.social
· Jun 17
Laura Eme
@lauraeme.bsky.social
· Jun 17
Laura Eme
@lauraeme.bsky.social
· Jun 17
Laura Eme
@lauraeme.bsky.social
· Jun 17
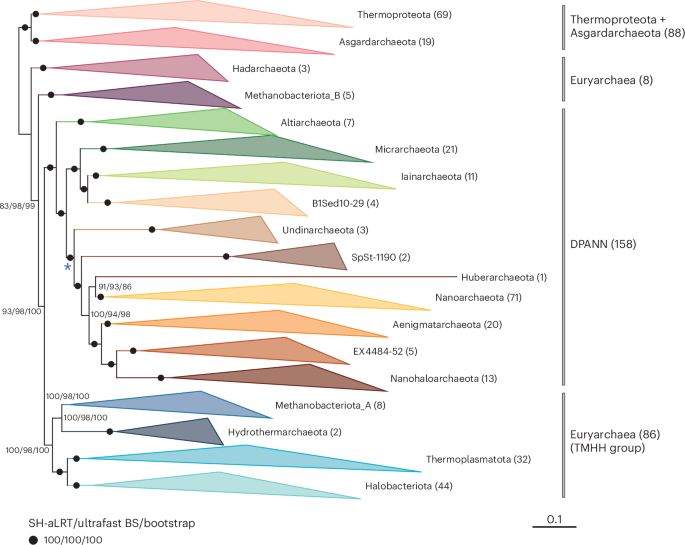
Phylogenomic analyses indicate the archaeal superphylum DPANN originated from free-living euryarchaeal-like ancestors - Nature Microbiology
Phylogenetic reconstructions with conserved protein markers from the 11 known DPANN phyla reveal their monophyletic placement within the Euryarchaeota.
www.nature.com
Laura Eme
@lauraeme.bsky.social
· May 25
Reposted by Laura Eme
Laura Eme
@lauraeme.bsky.social
· May 12
Reposted by Laura Eme
Jason Stajich
@hyphaltip.bsky.social
· May 9

Spotlight on DSMZ: Germany’s Leading Microbial Culture Collection - United States Culture Collection Network
USCCN Project Manager Dusti Gallagher and Steering Committee member Matthew Ryan (CABI) participated in the MICROBE project (The MICRObiome Biobanking (RI) Enabler) 2nd Annual Consortium Meeting held ...
usccn.org
Reposted by Laura Eme