Posts
Media
Videos
Starter Packs
Reposted by Laura Markey
Laura Markey
@lauramarkey.bsky.social
· Jun 25
Laura Markey
@lauramarkey.bsky.social
· Jun 25
Laura Markey
@lauramarkey.bsky.social
· Jun 25

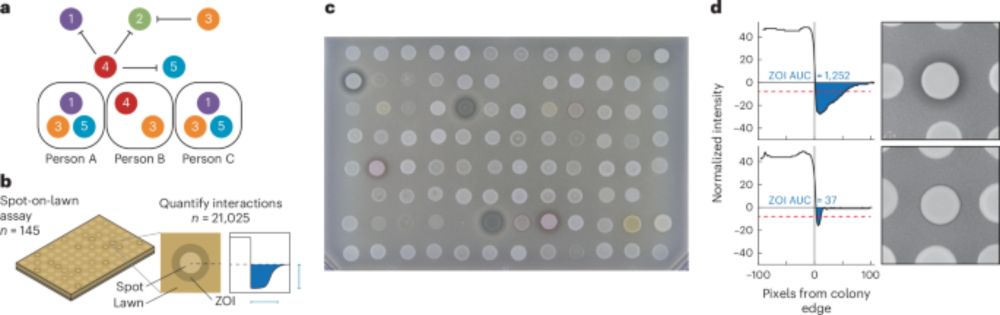
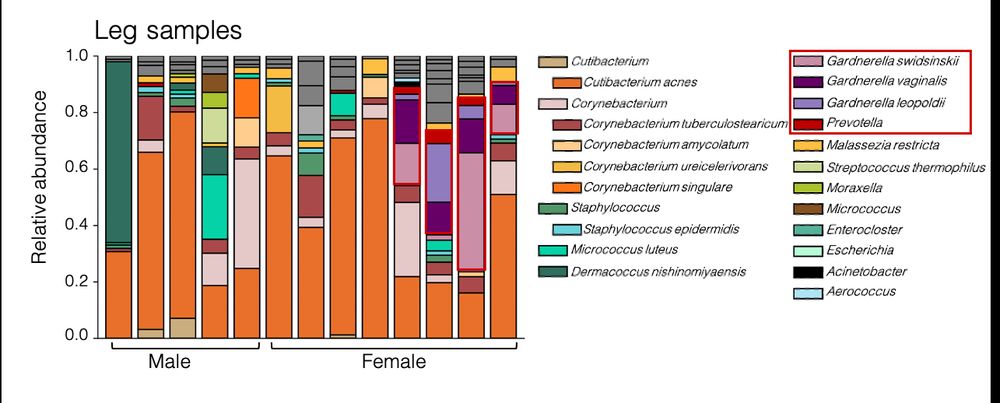
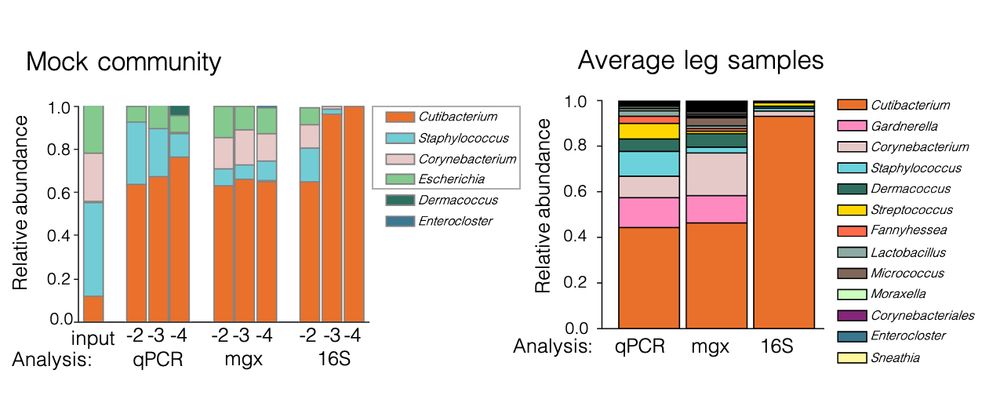
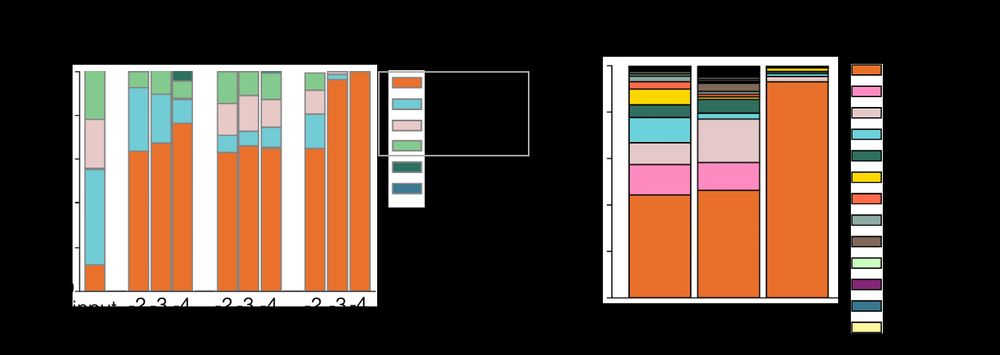
Microbiome diversity of low biomass skin sites is captured by metagenomics but not 16S amplicon sequencing
Established workflows for microbiome analysis work well for high microbial biomass samples, like stool, but often fail to accurately define microbial communities when applied to low microbial biomass ...
www.biorxiv.org
Laura Markey
@lauramarkey.bsky.social
· Feb 28
Laura Markey
@lauramarkey.bsky.social
· Feb 27
Reposted by Laura Markey
Evan Qu
@quevan.bsky.social
· Feb 11

Intraspecies associations from strain-rich metagenome samples
Genetically distinct strains of a species can vary widely in phenotype, reducing the utility of species-resolved microbiome measurements for detecting associations with health or disease. While metage...
www.biorxiv.org
Laura Markey
@lauramarkey.bsky.social
· Jan 24
Reposted by Laura Markey
Reposted by Laura Markey
Laura Markey
@lauramarkey.bsky.social
· Dec 6
Laura Markey
@lauramarkey.bsky.social
· Dec 6
Laura Markey
@lauramarkey.bsky.social
· Dec 5
Laura Markey
@lauramarkey.bsky.social
· Dec 5