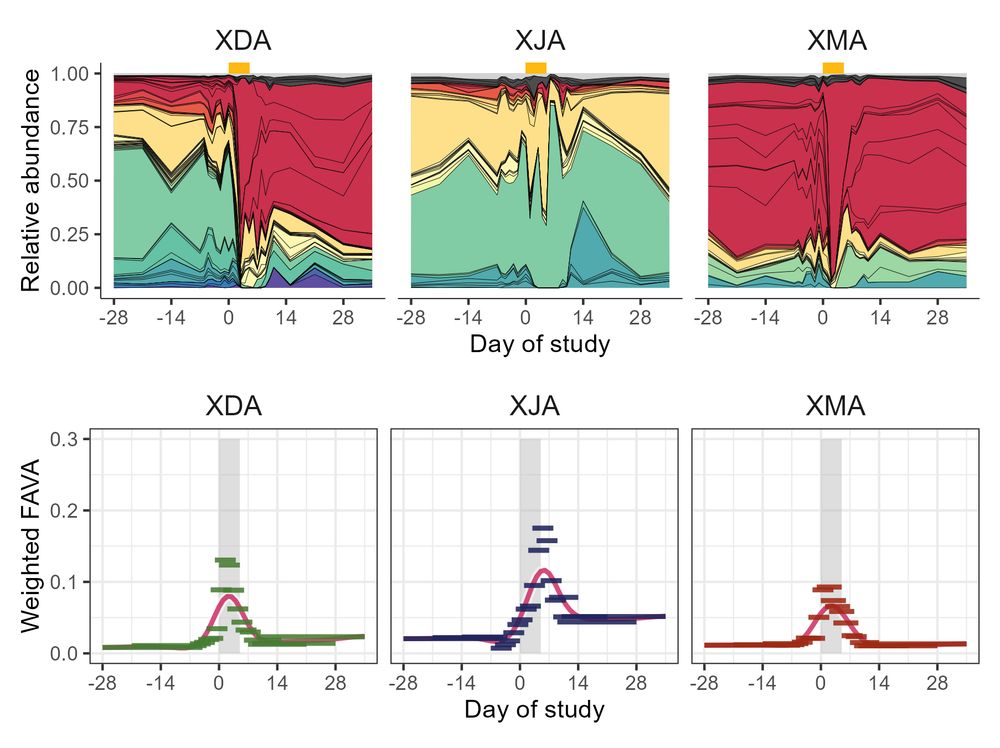
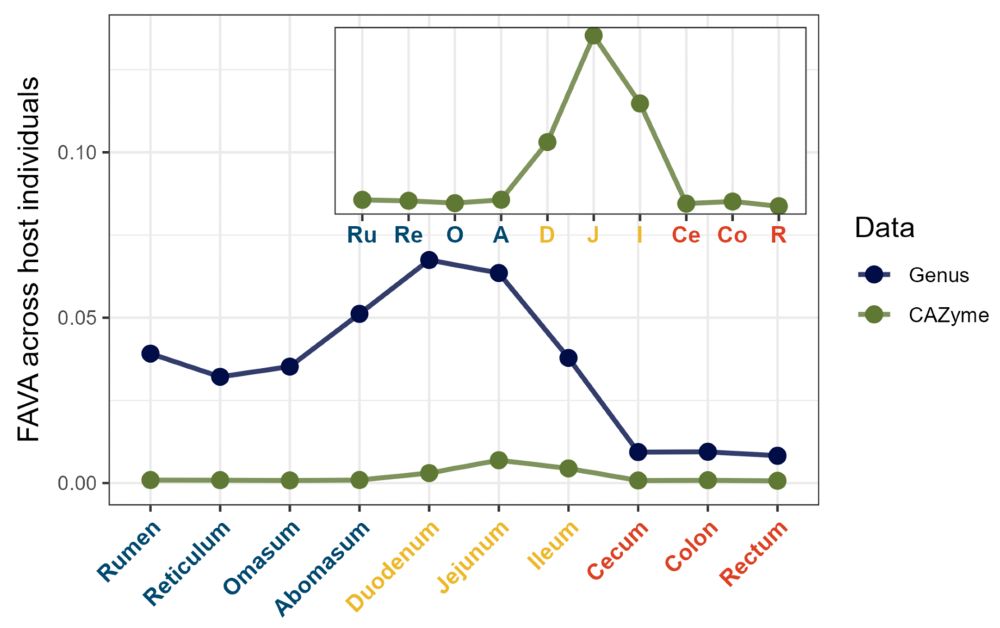
Maike Morrison
@maikemorrison.bsky.social
650 followers
160 following
31 posts
Building mathematical tools to answer biological questions
Ecology & evolution, sometimes microbiomes & cancer
Omidyar Fellow @SFIscience.bsky.social
Former PhD student with Noah Rosenberg, Stanford University
MaikeMorrison.com
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Maike Morrison
Ben Moran
@ben-moran.bsky.social
· May 1

Increased rates of hybridization in swordtail fish are associated with water pollution
Biodiversity loss can occur when disturbance compromises the reproductive barriers between species, causing them to collapse into a single population through hybridization. Recent research has documen...
www.biorxiv.org
Reposted by Maike Morrison
Maike Morrison
@maikemorrison.bsky.social
· Mar 17

Variability of mutational signatures is a footprint of carcinogens
Understanding the genomic impact of carcinogens is fundamental to cancer biology and prevention. However, recent coordinated efforts to detect such fingerprints have been largely unsuccessful, challen...
doi.org
Maike Morrison
@maikemorrison.bsky.social
· Mar 14
Maike Morrison
@maikemorrison.bsky.social
· Mar 14
Maike Morrison
@maikemorrison.bsky.social
· Mar 14
Maike Morrison
@maikemorrison.bsky.social
· Mar 14
Maike Morrison
@maikemorrison.bsky.social
· Mar 14
Maike Morrison
@maikemorrison.bsky.social
· Mar 14
Reposted by Maike Morrison
Maike Morrison
@maikemorrison.bsky.social
· Mar 12
Maike Morrison
@maikemorrison.bsky.social
· Jul 23