Marie Winz
@mariewinz.bsky.social
170 followers
260 following
13 posts
RNA biologist at Johannes Gutenberg University Mainz, Germany: co-translational quality control - RNA modification - and beyond.
Lab url: ak-winz.pharmazie.uni-mainz.de
Posts
Media
Videos
Starter Packs
Reposted by Marie Winz
Marie Winz
@mariewinz.bsky.social
· Sep 5
Reposted by Marie Winz
Eva Kowalinski
@kowaeva.bsky.social
· Jul 17

Postdoc in Structural Biology of RNA processing complexes
Postdoctoral positions in structural biology of macromolecular complexes are available in the laboratory of Dr. Eva Kowalinski at the EMBL Grenoble, France. We are looking for highly motivated and amb...
tinyurl.com
Marie Winz
@mariewinz.bsky.social
· May 23
Marie Winz
@mariewinz.bsky.social
· Apr 17
Reposted by Marie Winz
Molecular Cell
@cp-molcell.bsky.social
· Mar 26
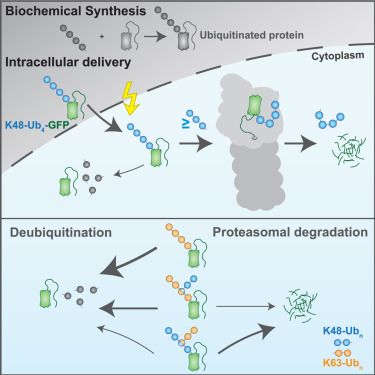
UbiREAD deciphers proteasomal degradation code of homotypic and branched K48 and K63 ubiquitin chains
Ubiquitin chains determine the fates of their modified proteins, including proteasomal degradation. Kiss et al. present UbiREAD, a technology to monitor cellular degradation and deubiquitination at high temporal resolution after intracellular delivery of ubiquitinated proteins. This reveals a degradation code for ubiquitin chains varying by linkage, length, and topology.
dlvr.it
Marie Winz
@mariewinz.bsky.social
· Feb 25
Marie Winz
@mariewinz.bsky.social
· Feb 25
Marie Winz
@mariewinz.bsky.social
· Feb 25
Marie Winz
@mariewinz.bsky.social
· Feb 25