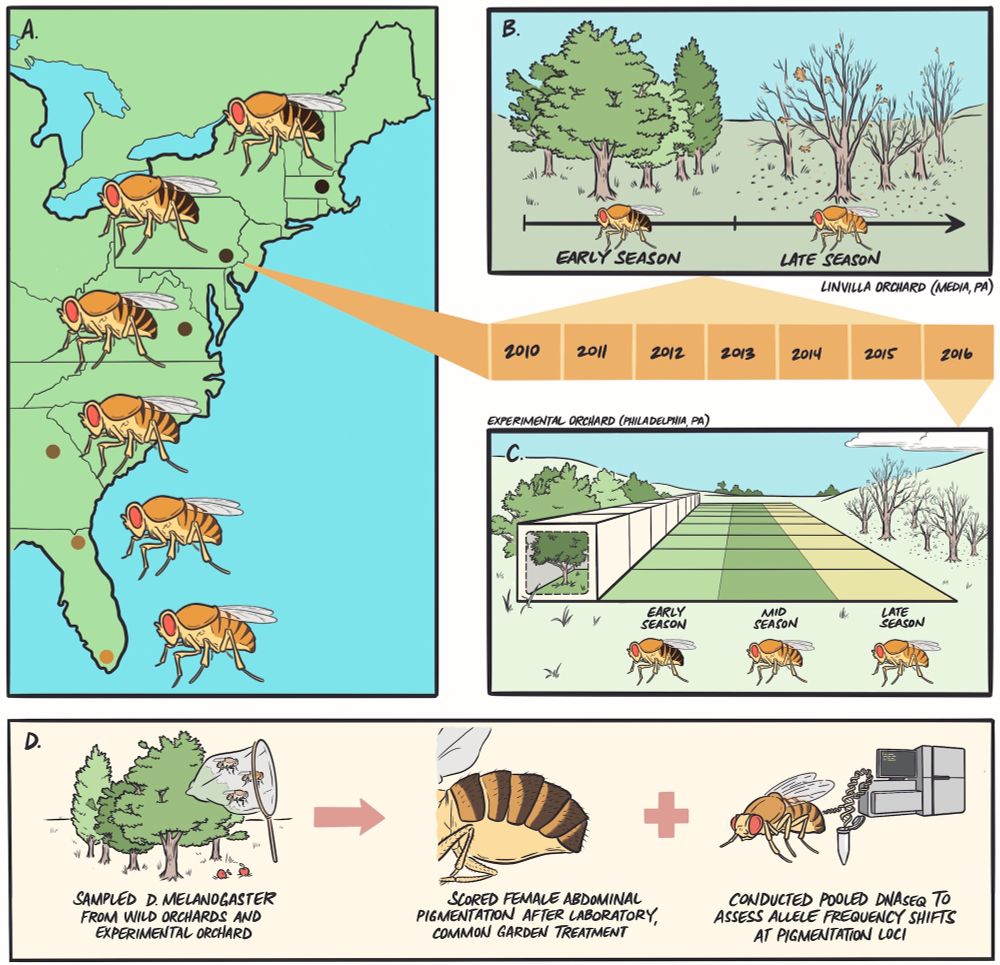
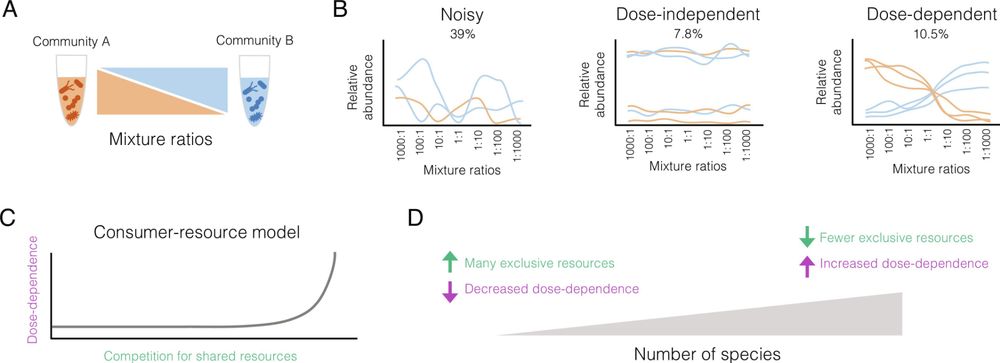
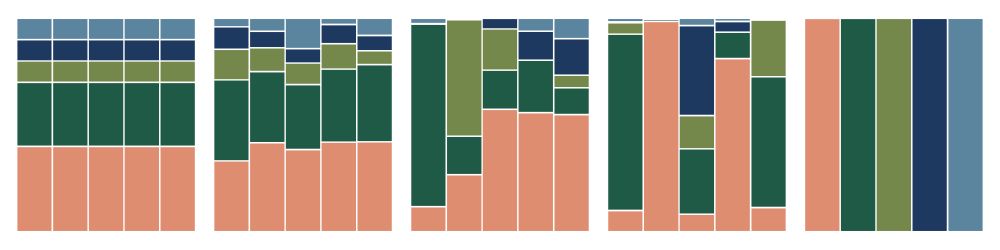
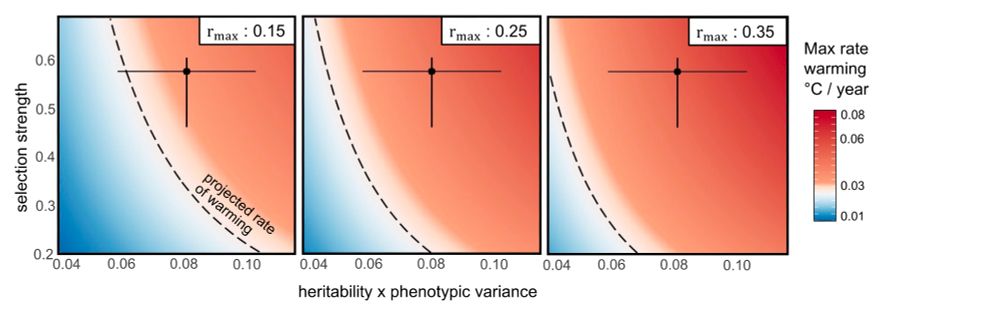
Mark Bitter
@markcbitter.bsky.social
230 followers
270 following
61 posts
Postdoc at Stanford University.
Interested in pop. gen., adaptation, and a wholesome scientific community.
he/him
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Skyler Berardi
@skylerberardi.bsky.social
· Jul 31
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Manuel Razo
@mrazo.bsky.social
· May 15
Reposted by Mark Bitter
Ben Moran
@ben-moran.bsky.social
· May 1

Increased rates of hybridization in swordtail fish are associated with water pollution
Biodiversity loss can occur when disturbance compromises the reproductive barriers between species, causing them to collapse into a single population through hybridization. Recent research has documen...
www.biorxiv.org
Reposted by Mark Bitter
James Hemker
@jahemker.bsky.social
· Apr 25

Manual validation finds only ultra-long long-read sequencing enables faithful, population-level structural variant calling in Drosophila melanogaster euchromatin
The increasing accessibility of long-read sequencing and the rapid development of automated variant callers are promoting the generation of population-level structural variation data. However, the eff...
doi.org
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Reposted by Mark Bitter
Marianna Karageorgi
@mkarag.bsky.social
· Jan 22

Dominance reversal maintains large-effect resistance polymorphism in temporally varying environments
A central challenge in evolutionary biology is to uncover mechanisms maintaining functional genetic variation1. Theory suggests that dominance reversal, whereby alleles subject to fluctuating selectio...
www.biorxiv.org
Reposted by Mark Bitter
Mark Bitter
@markcbitter.bsky.social
· Jan 21