Mark Robinson
@markrobinsonca.bsky.social
1.4K followers
300 following
13 posts
statistical bioinformatics; canadian / swiss; #methodsmatter #rstats; open science advocate; grumpy towards prestige worshippers, excessive admin and bros; will sacrifice sleep for a sunrise.
https://robinsonlabuzh.github.io/
Posts
Media
Videos
Starter Packs
Reposted by Mark Robinson
Reposted by Mark Robinson
Reposted by Mark Robinson
Reposted by Mark Robinson
Reposted by Mark Robinson
Martin Emons
@martinemons.bsky.social
· Jun 27

Harnessing the Potential of Spatial Statistics for Spatial Omics Data with pasta
Spatial omics assays allow for the molecular characterisation of cells in their spatial context. Notably, the two main technological streams, imaging-based and high-throughput sequencing-based, can gi...
arxiv.org
Reposted by Mark Robinson
Nav
@naveed-ishaque.bsky.social
· Jul 2

Beyond benchmarking: an expert-guided consensus approach to spatially aware clustering
Spatial omics technologies have revolutionized the study of tissue architecture and cellular heterogeneity by integrating molecular profiles with spatial localization. In spatially resolved transcript...
www.biorxiv.org
Reposted by Mark Robinson
Reposted by Mark Robinson
Reposted by Mark Robinson
Mark Robinson
@markrobinsonca.bsky.social
· Apr 29
Mark Robinson
@markrobinsonca.bsky.social
· Mar 24

Many Analysts, One Data Set: Making Transparent How Variations in Analytic Choices Affect Results - R. Silberzahn, E. L. Uhlmann, D. P. Martin, P. Anselmi, F. Aust, E. Awtrey, Š. Bahník, F. Bai, C. Ba...
Twenty-nine teams involving 61 analysts used the same data set to address the same research question: whether soccer referees are more likely to give red cards ...
journals.sagepub.com
Reposted by Mark Robinson
Reposted by Mark Robinson
Reposted by Mark Robinson
Reposted by Mark Robinson
Neuron
@cp-neuron.bsky.social
· Dec 20
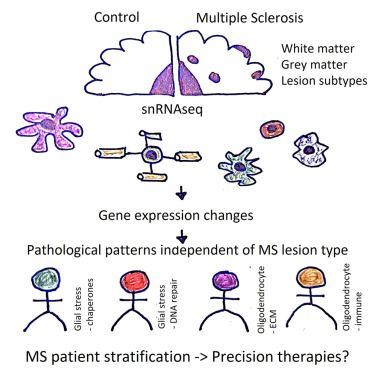
snRNA-seq stratifies multiple sclerosis patients into distinct white matter glial responses
Macnair and Calini et al. analyze 632,000 snRNA-seq profiles from multiple sclerosis and control brain samples, identifying distinct cellular responses in white and gray matter. They stratify MS patients into groups based on patterns of gene expression in…
dlvr.it
Siyuan Luo
@siyuanluo.bsky.social
· Dec 5

On metrics for subpopulation detection in single-cell and spatial omics data
Benchmarks are crucial to understanding the strengths and weaknesses of the growing number of tools for single-cell and spatial omics analysis. A key task is to distinguish subpopulations within compl...
doi.org