Nav
@naveed-ishaque.bsky.social
910 followers
1.2K following
25 posts
Bioinformatics research group leader at the Berlin Institute of Health, ELIXIR and de.NBI. Main focus: cancer, immunology, placenta, and all things omics... especially spatial omics.
Posts
Media
Videos
Starter Packs
Nav
@naveed-ishaque.bsky.social
· Jul 4

Sainsc: A Computational Tool for Segmentation‐Free Analysis of In Situ Capture Data (Small Methods 5/2025)
Segmentation-Free Analyses Sainsc: a new tool for efficient whole organism spatial transcriptomics data analysis at the nanometre scale. Shown is a blended composite spatial map of total gene express...
onlinelibrary.wiley.com
Nav
@naveed-ishaque.bsky.social
· Jul 3
Nav
@naveed-ishaque.bsky.social
· Jul 3
Nav
@naveed-ishaque.bsky.social
· Jul 2
Nav
@naveed-ishaque.bsky.social
· Jul 2

Beyond benchmarking: an expert-guided consensus approach to spatially aware clustering
Spatial omics technologies have revolutionized the study of tissue architecture and cellular heterogeneity by integrating molecular profiles with spatial localization. In spatially resolved transcript...
www.biorxiv.org
Nav
@naveed-ishaque.bsky.social
· Mar 17

Segger: Fast and accurate cell segmentation of imaging-based spatial transcriptomics data
The accurate assignment of transcripts to their cells of origin remains the Achilles heel of imaging-based spatial transcriptomics, despite being critical for nearly all downstream analyses. Current c...
www.biorxiv.org
Nav
@naveed-ishaque.bsky.social
· Feb 8

2D, or not 2D? Investigating Vertical Signal Integrity of Tissue Slices
Imaging-based spatially resolved transcriptomics can localise transcripts within cells in 3D. Cell segmentation precedes assignment of transcripts to cells and annotation of cell function. However, ce...
t.ly
Nav
@naveed-ishaque.bsky.social
· Feb 8
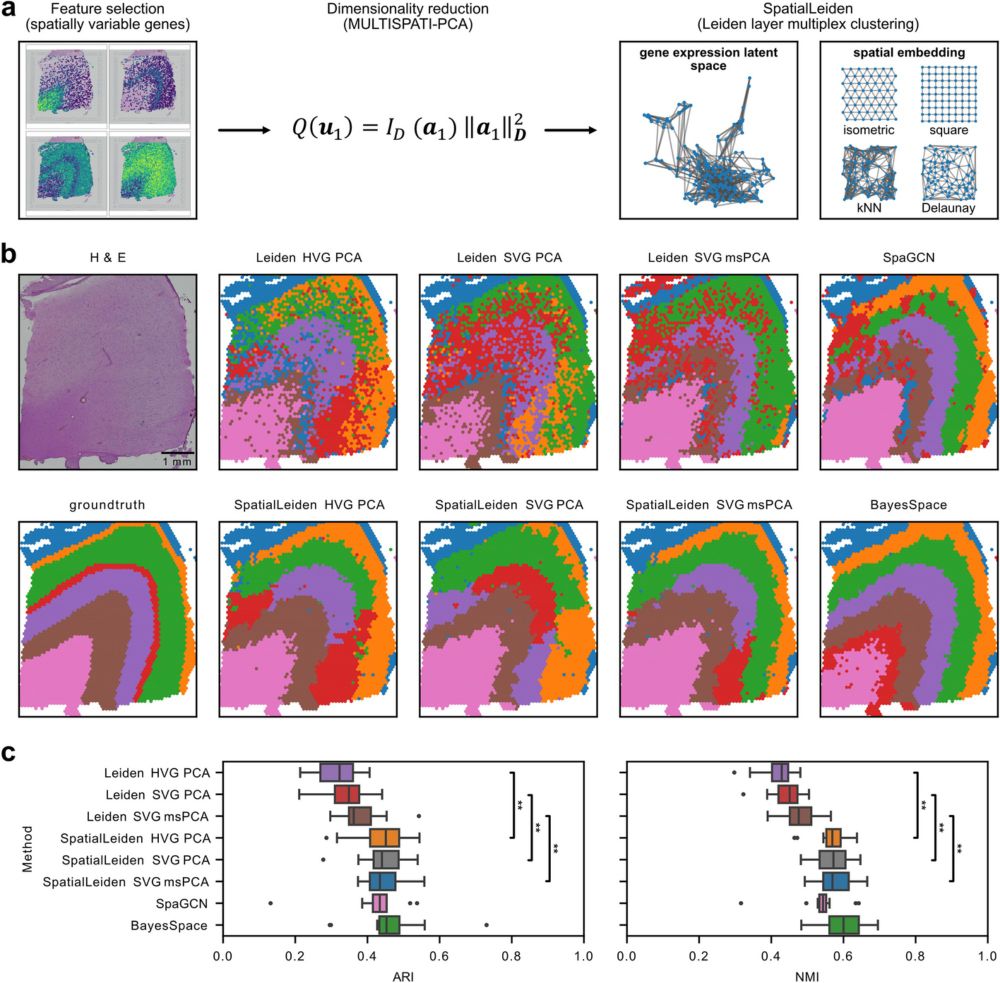
SpatialLeiden: spatially aware Leiden clustering - Genome Biology
Clustering can identify the natural structure that is inherent to measured data. For single-cell omics, clustering finds cells with similar molecular phenotype after which cell types are annotated. Le...
t.ly
Nav
@naveed-ishaque.bsky.social
· Jan 8
Nav
@naveed-ishaque.bsky.social
· Jan 8

BIH PhD Program - Call for PhD candidates 2025 - News - BIH at Charité
The BIH PhD program aims to promote interdisciplinarity and support young translational talents. The program co-funds PhD positions for excellent translational projects that fit one of the four BIH re...
www.bihealth.org
Reposted by Nav
Nav
@naveed-ishaque.bsky.social
· Dec 19
Nav
@naveed-ishaque.bsky.social
· Dec 17
Nav
@naveed-ishaque.bsky.social
· Dec 3
Nav
@naveed-ishaque.bsky.social
· Dec 3

SpatialLeiden - Spatially-aware Leiden clustering
Clustering can identify the natural structure that is inherent to measured data. For single-cell omics, clustering finds cells with similar molecular phenotype after which cell types are annotated. Le...
www.biorxiv.org
Reposted by Nav