Martina Braun
@martinabraun.bsky.social
960 followers
300 following
10 posts
PhD Candidate @CRG.eu with @larsplus.bsky.social | Computational Biology & Single Cell Epigenetics
Posts
Media
Videos
Starter Packs
Pinned
Martina Braun
@martinabraun.bsky.social
· May 21
Lars Velten
@larsplus.bsky.social
· May 21
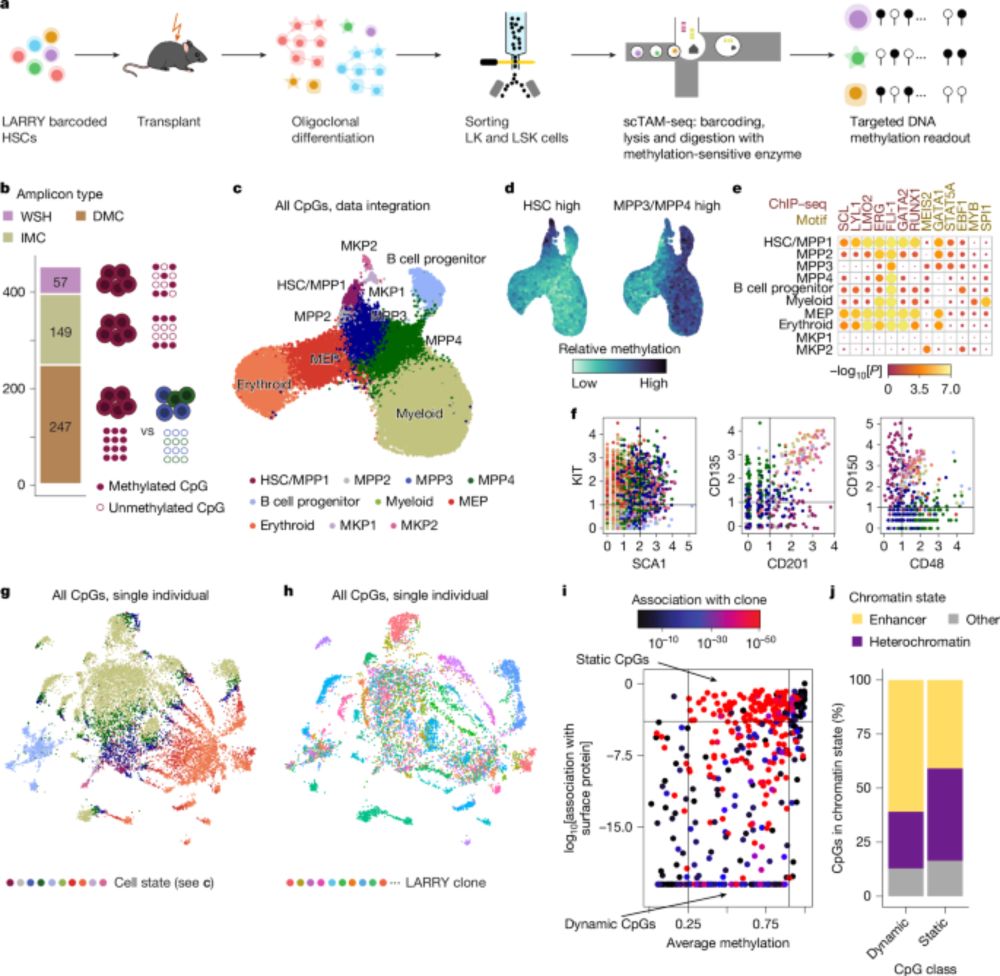
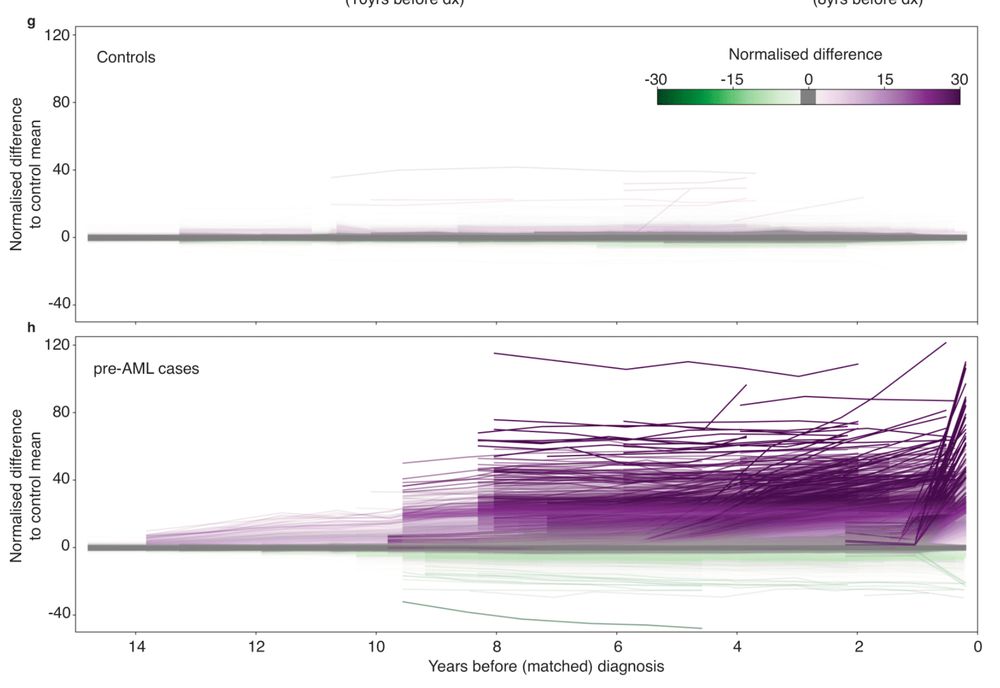
Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing th...
doi.org
Reposted by Martina Braun
Reposted by Martina Braun
Reposted by Martina Braun
Reposted by Martina Braun
Lars Velten
@larsplus.bsky.social
· May 31
„Blutbarcodes“ könnten Alterung verlangsamen
Auch das Blut altert: Ab 50 Jahren verringert sich die Vielfalt der Stammzellen, die Blut produzieren. Ein Forschungsteam hat nun ein „Barcodesystem“ entwickelt, das die verschiedenen Blutstammzellen ...
science.orf.at
Reposted by Martina Braun
Charlie Pugh
@cwjpugh.bsky.social
· May 26

From Likelihood to Fitness: Improving Variant Effect Prediction in Protein and Genome Language Models
Generative models trained on natural sequences are increasingly used to predict the effects of genetic variation, enabling progress in therapeutic design, disease risk prediction, and synthetic biolog...
www.biorxiv.org
Reposted by Martina Braun
Nature
@nature.com
· May 21
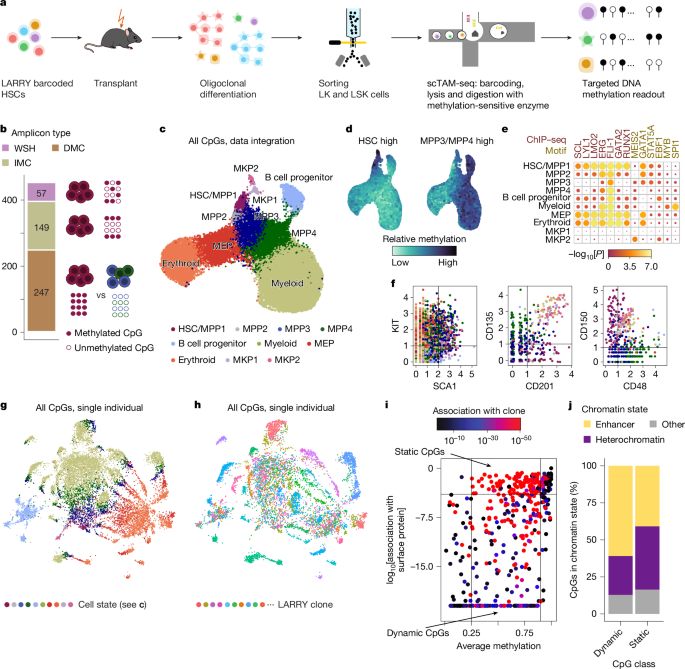
Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing through cellular differentiation at scale.
go.nature.com
Reposted by Martina Braun
Lars Velten
@larsplus.bsky.social
· May 21

Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing th...
doi.org
Martina Braun
@martinabraun.bsky.social
· May 21
Martina Braun
@martinabraun.bsky.social
· May 21
Lars Velten
@larsplus.bsky.social
· May 21

Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing th...
doi.org
Reposted by Martina Braun
Michael Scherer
@scherermich.bsky.social
· May 21
Lars Velten
@larsplus.bsky.social
· May 21

Clonal tracing with somatic epimutations reveals dynamics of blood ageing - Nature
The discovery that DNA methylation of different CpG sites can serve as digital barcodes of clonal identity led to the development of EPI-Clone, an algorithm that enables single-cell lineage tracing th...
doi.org
Reposted by Martina Braun
Michael Scherer
@scherermich.bsky.social
· May 21
Reposted by Martina Braun
Reposted by Martina Braun
Lars Velten
@larsplus.bsky.social
· May 8

Design principles of cell-state-specific enhancers in hematopoiesis
Screen of minimalistic enhancers in blood progenitor cells demonstrates widespread
dual activator-repressor function of transcription factors (TFs) and enables the model-guided
design of cell-state-sp...
www.cell.com
Reposted by Martina Braun
Reposted by Martina Braun
Martina Braun
@martinabraun.bsky.social
· Nov 25
Martina Braun
@martinabraun.bsky.social
· Nov 23
Martina Braun
@martinabraun.bsky.social
· Nov 22
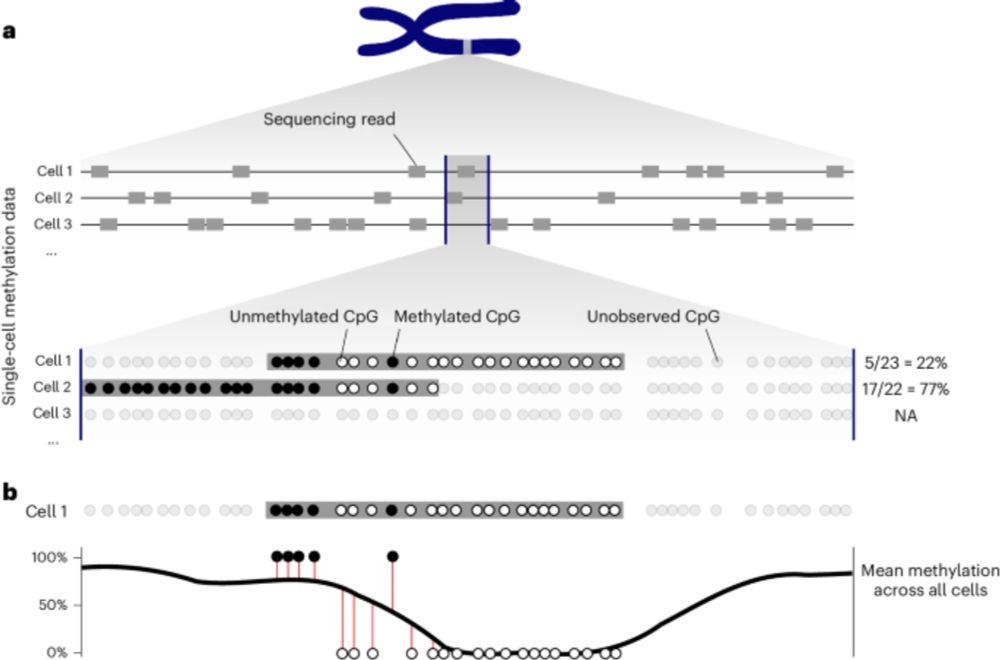
Analyzing single-cell bisulfite sequencing data with MethSCAn - Nature Methods
This work highlights the technical issues in previous approaches and introduces a preprocessing approach along with a software package, MethSCAn, for single-cell bisulfite sequencing data analysis.
www.nature.com