Matt Olm
@mattolm.bsky.social
1.9K followers
200 following
28 posts
Assistant Professor at CU Boulder
Studying the human microbiome
https://www.colorado.edu/lab/olm/
Posts
Media
Videos
Starter Packs
Matt Olm
@mattolm.bsky.social
· 28d
Matt Olm
@mattolm.bsky.social
· Aug 30
Matt Olm
@mattolm.bsky.social
· Jul 17
Reposted by Matt Olm
Tom Booth
@boothicus.bsky.social
· Jul 9

The spatiotemporal distribution of human pathogens in ancient Eurasia - Nature
Screening shotgun-sequencing data from ancient humans covering 37,000 years of Eurasian history uncovers the widespread presence of ancient bacterial, viral and parasite DNA and zoonotic pathogens coi...
www.nature.com
Reposted by Matt Olm
Reposted by Matt Olm
Reposted by Matt Olm
Trenton Dailey-Chwalibóg
@trenton.phd
· Mar 25
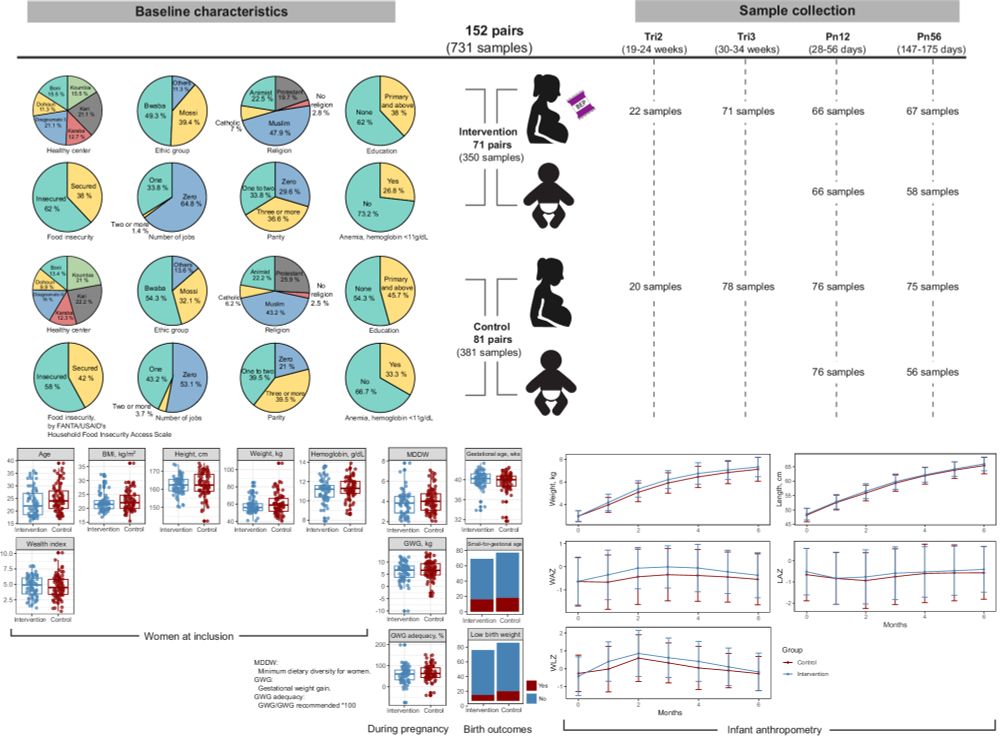
Maternal balanced energy-protein supplementation reshapes the maternal gut microbiome and enhances carbohydrate metabolism in infants: a randomized controlled trial
Nature Communications - Here, in a sub-study of the MISAME-III randomized controlled trial, the authors show that maternal balanced energy-protein supplementation during pregnancy and lactation...
rdcu.be
Reposted by Matt Olm
Reposted by Matt Olm
Reposted by Matt Olm
Reposted by Matt Olm
Matt Olm
@mattolm.bsky.social
· Jan 5
Matt Olm
@mattolm.bsky.social
· Jan 3
Matt Olm
@mattolm.bsky.social
· Jan 3
Matt Olm
@mattolm.bsky.social
· Jan 3
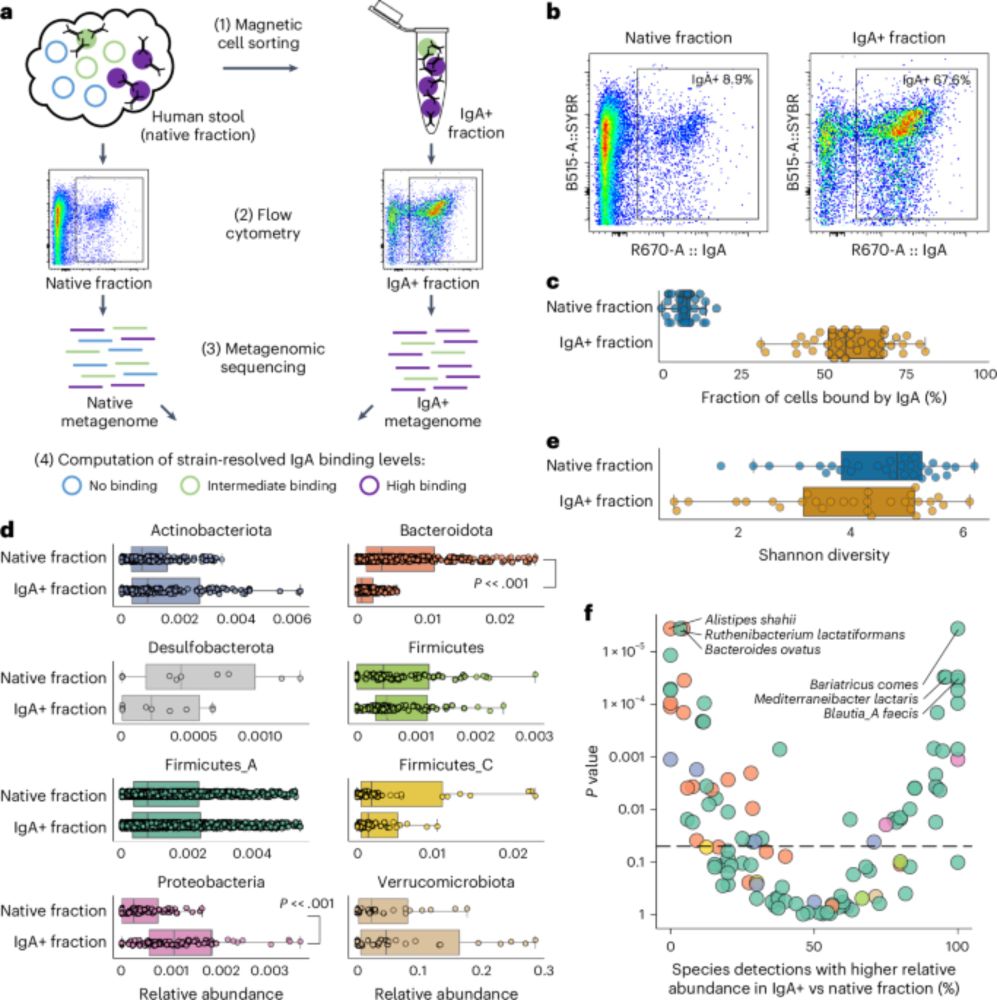
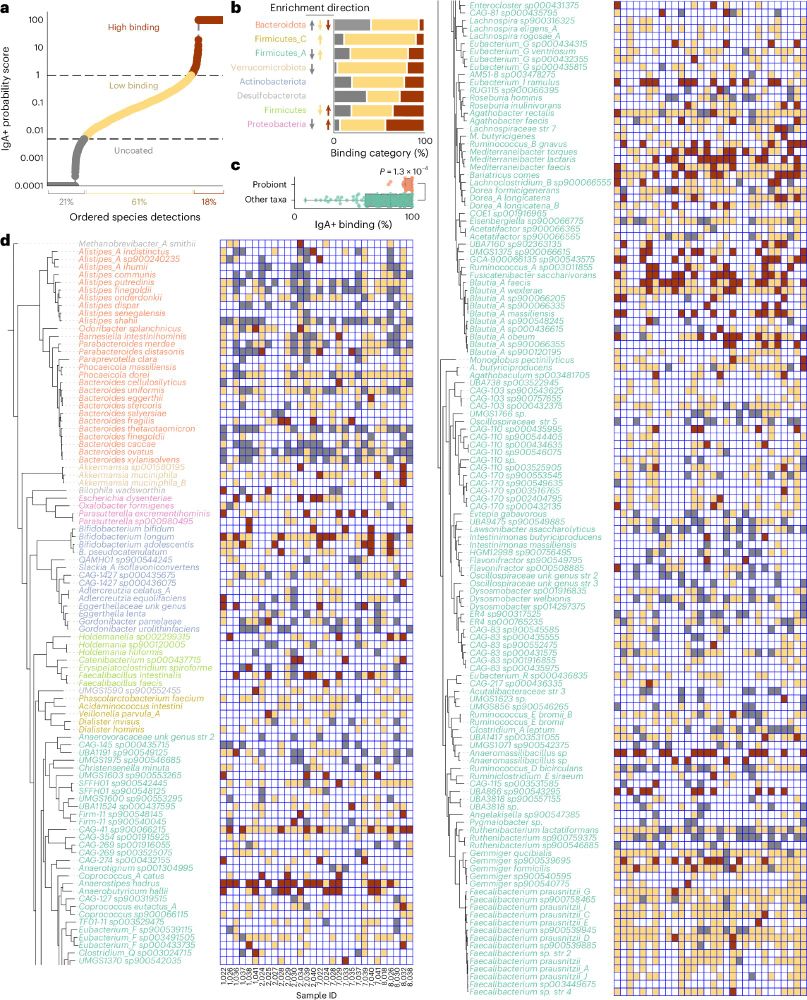
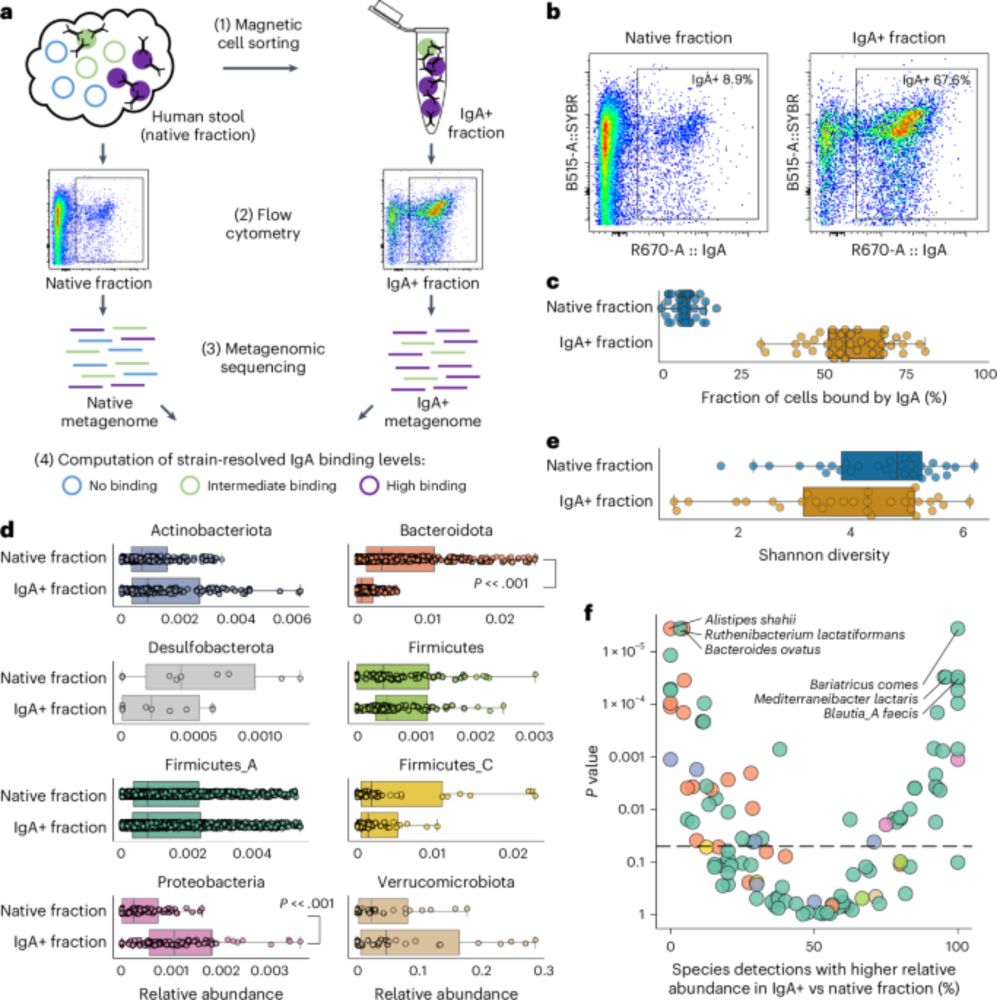
Metagenomic immunoglobulin sequencing reveals IgA coating of microbial strains in the healthy human gut - Nature Microbiology
Metagenomic immunoglobulin sequencing (MIg-seq) uncovered patterns of IgA antibody binding of bacterial strains in the healthy human gut microbiome.
doi.org
Matt Olm
@mattolm.bsky.social
· Jan 3