Matt Bogyo
@mbogyo.bsky.social
610 followers
360 following
44 posts
Chemical biologist at Stanford with a love for covalent binding molecules and hydrolases.
Posts
Media
Videos
Starter Packs
Matt Bogyo
@mbogyo.bsky.social
· Aug 29

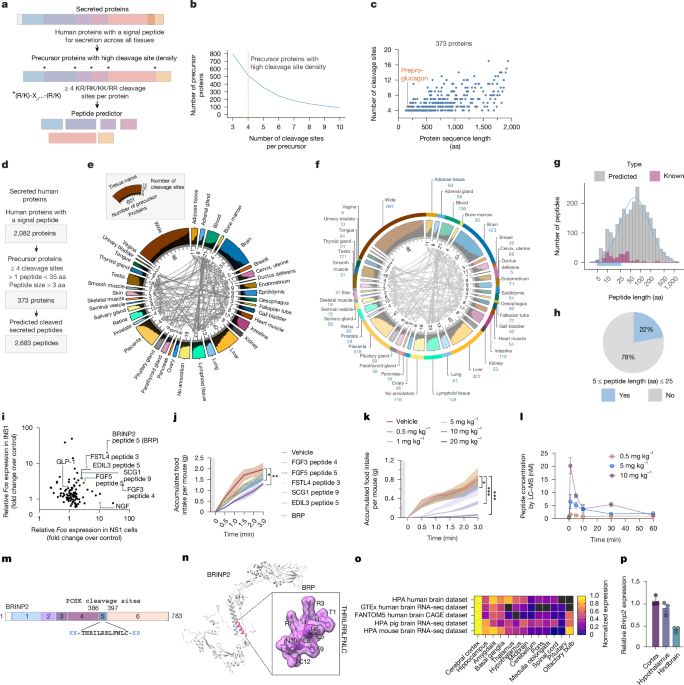
A Two-Step Synthesis of Covalent Genetically-Encoded Libraries of Peptide-Derived Macrocycles (cGELs) enables use of electrophiles with diverse reactivity
Genetically-encoded libraries of peptide-derived macrocycles containing electrophile "warheads" (cGELs) can be used to identify potent and selective covalent ligands for protein targets. Such cGELs ar...
www.biorxiv.org
Matt Bogyo
@mbogyo.bsky.social
· Jul 19

Macrocyclic Phage Display for Identification of Selective Protease Substrates
Traditional methods for identifying selective protease substrates have primarily relied on synthetic libraries of linear peptides, which offer limited sequence and structural diversity. Here, we present an approach that leverages phage display technology to screen large libraries of chemically modified cyclic peptides, enabling the identification of highly selective substrates for a protease of interest. Our method uses a reactive chemical linker to cyclize peptides on the phage surface, while simultaneously incorporating an affinity tag and a fluorescent reporter. The affinity tag enables capture of the phage library and subsequent release of phages expressing optimal substrates upon incubation with a protease of interest. The addition of a turn-on fluorescent reporter allows direct quantification of cleavage efficiency throughout each selection round. The resulting identified substrates can then be chemically synthesized, optimized and validated using recombinant enzymes and cells. We demonstrate the utility of this approach using Fibroblast Activation Protein α (FAPα) and the related proline-specific protease, dipeptidyl peptidase-4 (DPP4), as targets. Phage selection and subsequent optimization identified substrates with selectivity for each target that have the potential to serve as valuable tools for applications in basic biology and fluorescence image-guided surgery (FIGS). Overall, our strategy provides a rapid and unbiased platform for effectively discovering highly selective, non-natural protease substrates, overcoming key limitations of existing methods.
pubs.acs.org
Matt Bogyo
@mbogyo.bsky.social
· May 31

A caspase-1-cathepsin AND-gate probe for selective imaging of inflammasome activation
Caspase-1 is a key mediator of the inflammasome pathway, which is associated with several inflammatory disorders including obesity, diabetes mellitus (DM), cardiovascular diseases (CVDs), cancers and ...
www.biorxiv.org
Matt Bogyo
@mbogyo.bsky.social
· May 30
Matt Bogyo
@mbogyo.bsky.social
· May 30
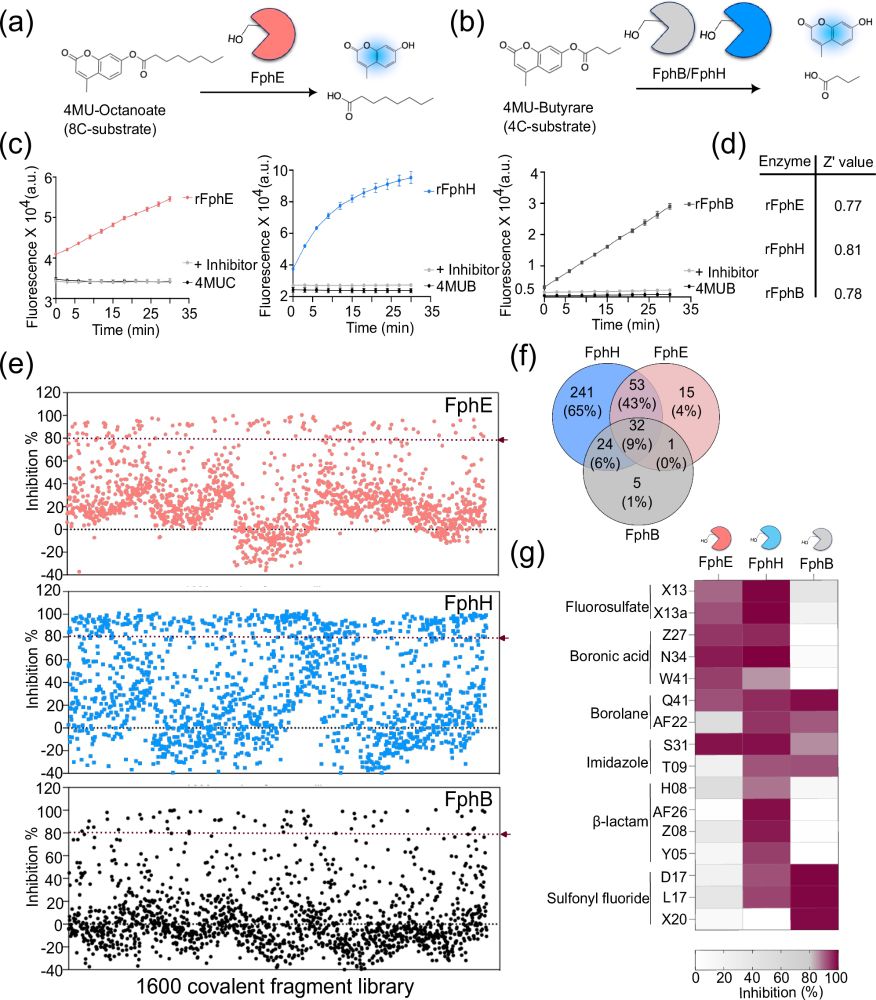
Identification of covalent inhibitors of Staphylococcus aureus serine hydrolases important for virulence and biofilm formation - Nature Communications
Staphylococcus aureus is a leading cause of bacteria-associated mortality worldwide. New tools are needed to both image and treat this pathogen. We previously identified a group of S. aureus serine hy...
link.springer.com
Reposted by Matt Bogyo
Reposted by Matt Bogyo
Anupam Patgiri
@apatgiri.bsky.social
· Mar 31

Engineered commensals for targeted nose-to-brain drug delivery
Shen et al. investigate the use of Lactobacillus plantarum, a commensal bacterial
strain, as a chassis for targeting the olfactory mucosa to facilitate precise nose-to-brain
delivery of therapeutic mo...
www.cell.com
Reposted by Matt Bogyo
Matt Bogyo
@mbogyo.bsky.social
· Mar 29
Matt Bogyo
@mbogyo.bsky.social
· Mar 26
Reposted by Matt Bogyo
Denis Wirtz
@deniswirtz.bsky.social
· Mar 13

NIH Grants Fueled $95 Billion In FY 2024 Economic Activity, Finds New Report
National Institutes of Health grants generated almost $95 billion in economic activity nationwide in FY 2024 according to a new report by United for Medical Research.
www.forbes.com
Reposted by Matt Bogyo