Josh Mendell
@mendell-lab.bsky.social
1K followers
590 following
35 posts
Scientist in the Department of Molecular Biology at UTSW/HHMI. We study post-transcriptional gene regulation & noncoding RNAs in normal physiology and disease.
https://labs.utsouthwestern.edu/mendell-lab
https://www.hhmi.org/scientists/joshua-t-mendell
Posts
Media
Videos
Starter Packs
Josh Mendell
@mendell-lab.bsky.social
· Jul 17
Reposted by Josh Mendell
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2
Josh Mendell
@mendell-lab.bsky.social
· Jul 2

Plagl1 and Lrrc58 control mammalian body size by triggering target-directed microRNA degradation of miR-322 and miR-503
Precise control of microRNA (miRNA) expression is critical during development. An important mechanism of miRNA regulation is target-directed microRNA degradation (TDMD), a pathway in which the binding...
www.biorxiv.org
Josh Mendell
@mendell-lab.bsky.social
· Apr 4
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
Josh Mendell
@mendell-lab.bsky.social
· Apr 3
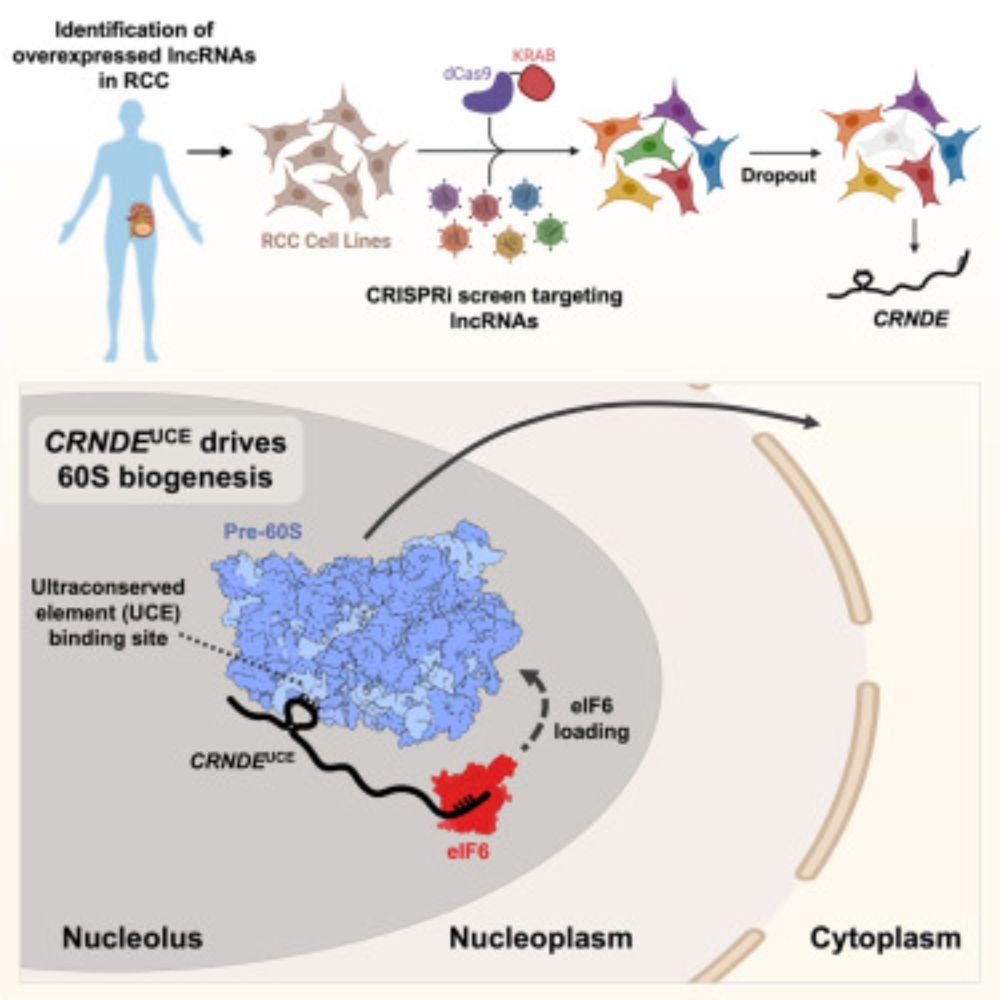
An ultraconserved snoRNA-like element in long noncoding RNA CRNDE promotes ribosome biogenesis and cell proliferation
Cancer cells frequently enhance ribosome production to support rapid growth. Here,
Lee et al. demonstrate that an isoform of lncRNA CRNDE containing an ultraconserved
element (CRNDEUCE) promotes 60S r...
www.cell.com
Josh Mendell
@mendell-lab.bsky.social
· Dec 19
Josh Mendell
@mendell-lab.bsky.social
· Nov 23



