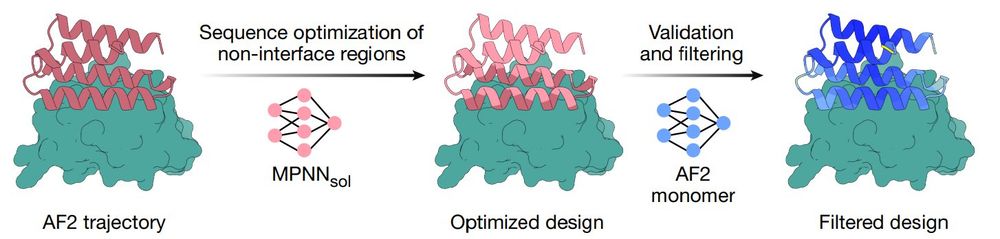
Protein evolution, metagenomics, AI/ML/DL
Website https://miangoaren.github.io/
youtu.be/_jDRr5BcTaY
Slides
drive.google.com/file/d/1i4QE...
English is available only via auto-translated subtitles

youtu.be/_jDRr5BcTaY
Slides
drive.google.com/file/d/1i4QE...
English is available only via auto-translated subtitles
youtu.be/qaypRS8SX5M
Slides
drive.google.com/file/d/1BfQd...
English is available only via auto-translated subtitles

youtu.be/qaypRS8SX5M
Slides
drive.google.com/file/d/1BfQd...
English is available only via auto-translated subtitles
youtu.be/cZs8XtVYa5A
Slides
drive.google.com/file/d/1TpPj...
English is available only via auto-translated subtitles

youtu.be/cZs8XtVYa5A
Slides
drive.google.com/file/d/1TpPj...
English is available only via auto-translated subtitles
youtu.be/gE6qXwpBP_s
Slides
drive.google.com/file/d/1F99V...
For now, the English version is only available through the automatic translation of the subtitles

youtu.be/gE6qXwpBP_s
Slides
drive.google.com/file/d/1F99V...
For now, the English version is only available through the automatic translation of the subtitles
youtu.be/vUpb6O6T2yQ
Slides
drive.google.com/file/d/1y2Vj...
For now, the English version is only available through the automatic translation of the subtitles.

youtu.be/vUpb6O6T2yQ
Slides
drive.google.com/file/d/1y2Vj...
For now, the English version is only available through the automatic translation of the subtitles.
youtu.be/pAgL7NsCUMU
Slides
drive.google.com/file/d/1cazt...
For now, the English version is only available through the automatic translation of the subtitles.

youtu.be/pAgL7NsCUMU
Slides
drive.google.com/file/d/1cazt...
For now, the English version is only available through the automatic translation of the subtitles.
youtu.be/Xx80O85-5rI
Slides
drive.google.com/file/d/1i-Jo...
For now, the English version is only available through the automatic translation of the subtitles.

youtu.be/Xx80O85-5rI
Slides
drive.google.com/file/d/1i-Jo...
For now, the English version is only available through the automatic translation of the subtitles.
youtu.be/uMkZzKbnoJI
Slides
drive.google.com/file/d/1uDwe...
For now, the English version is only available through the automatic translation of the subtitles.

youtu.be/uMkZzKbnoJI
Slides
drive.google.com/file/d/1uDwe...
For now, the English version is only available through the automatic translation of the subtitles.


Protein superfamilies and domain superfolds
pubmed.ncbi.nlm.nih.gov/7990952/

Protein superfamilies and domain superfolds
pubmed.ncbi.nlm.nih.gov/7990952/




Congrats to all the authors!
Congrats to all the authors!

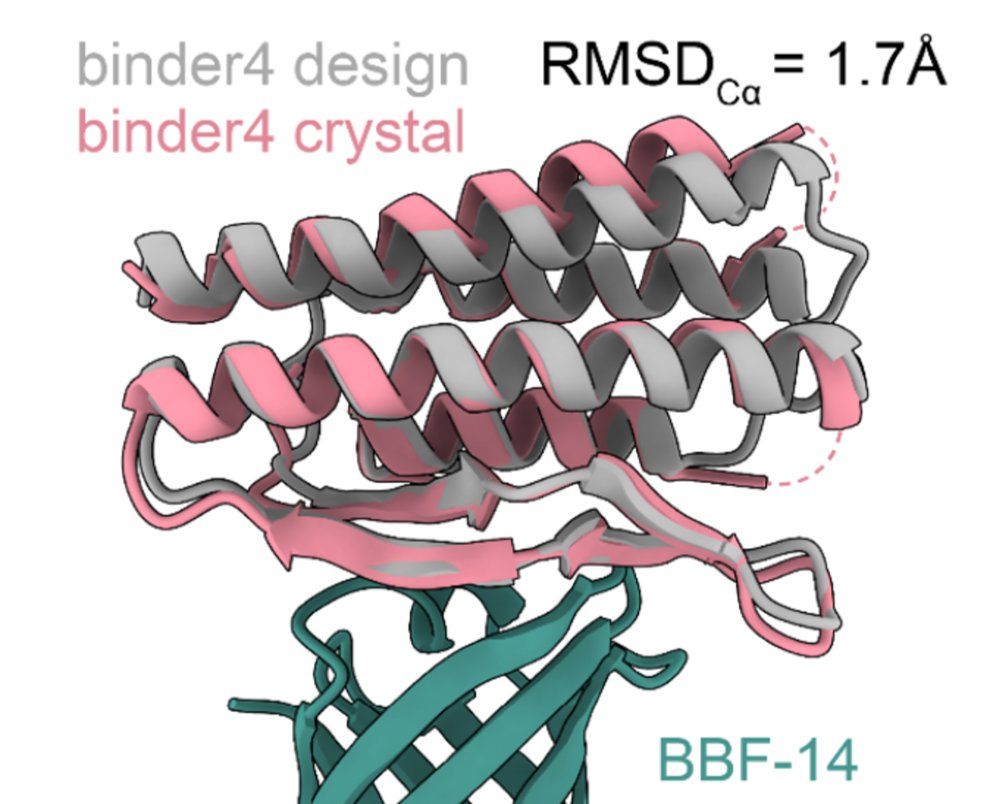
*proteins with no known binding sites
*membrane proteins , which are much harder than intra/extra-cellular proteins
*proteins lacking evolutionary information
*proteins that interact with DNA/RNA
*medically relevant proteins such as those causing allergies

*proteins with no known binding sites
*membrane proteins , which are much harder than intra/extra-cellular proteins
*proteins lacking evolutionary information
*proteins that interact with DNA/RNA
*medically relevant proteins such as those causing allergies