But that still doesn’t explain the complete lack of correlation with UPS2.
From my experience, iBAQ and MaxLFQ usually correlate well (R² ~0.78, non related example dataset shown), suggesting they track the same MS1 signal.

But that still doesn’t explain the complete lack of correlation with UPS2.
From my experience, iBAQ and MaxLFQ usually correlate well (R² ~0.78, non related example dataset shown), suggesting they track the same MS1 signal.

Anyone know what’s going on here? 🤔
#proteomics #bioinformatics @pwilmarth.bsky.social il

Anyone know what’s going on here? 🤔
#proteomics #bioinformatics @pwilmarth.bsky.social il
Many non-human proteomics studies still search against taxon-filtered FASTAs.
❌ Redundant sequences
❌ Inflated search space
✅ Reference proteomes cut redundancy, improve annotation, and make results comparable.
👉 Time to move beyond taxon filters. #proteomics #massspec #uniprot

Many non-human proteomics studies still search against taxon-filtered FASTAs.
❌ Redundant sequences
❌ Inflated search space
✅ Reference proteomes cut redundancy, improve annotation, and make results comparable.
👉 Time to move beyond taxon filters. #proteomics #massspec #uniprot


When you run proteomics on non-human species (mouse, rat, macaque, etc.) — which protein FASTA do you prefer?
Taxonomy-filtered UniProt (all entries)
Reference proteome (SwissProt+TrEMBL)
Ensembl/GENCODE
Something else?

When you run proteomics on non-human species (mouse, rat, macaque, etc.) — which protein FASTA do you prefer?
Taxonomy-filtered UniProt (all entries)
Reference proteome (SwissProt+TrEMBL)
Ensembl/GENCODE
Something else?
www.biorxiv.org/content/10.1...
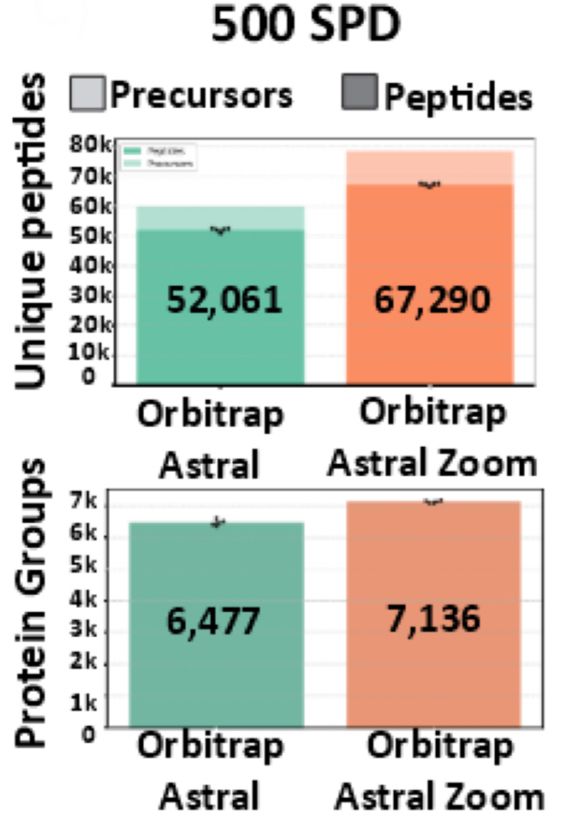
www.biorxiv.org/content/10.1...
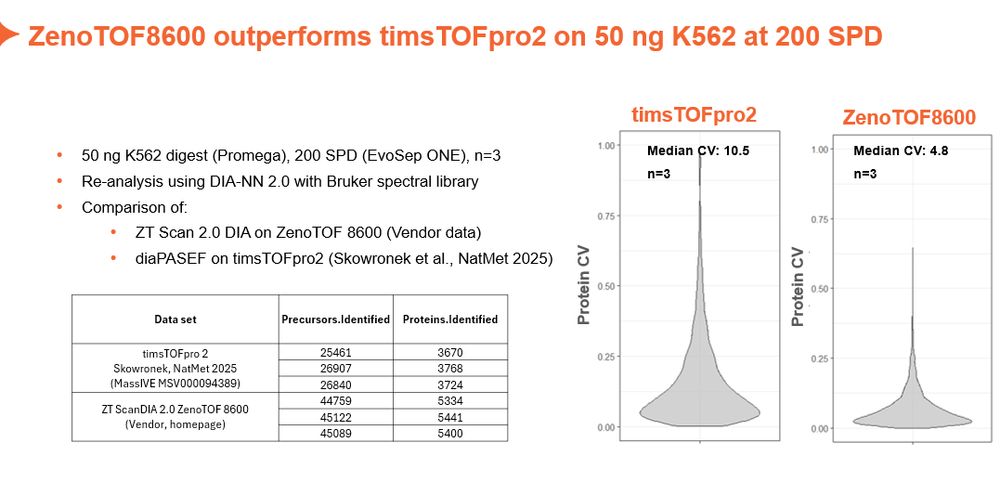
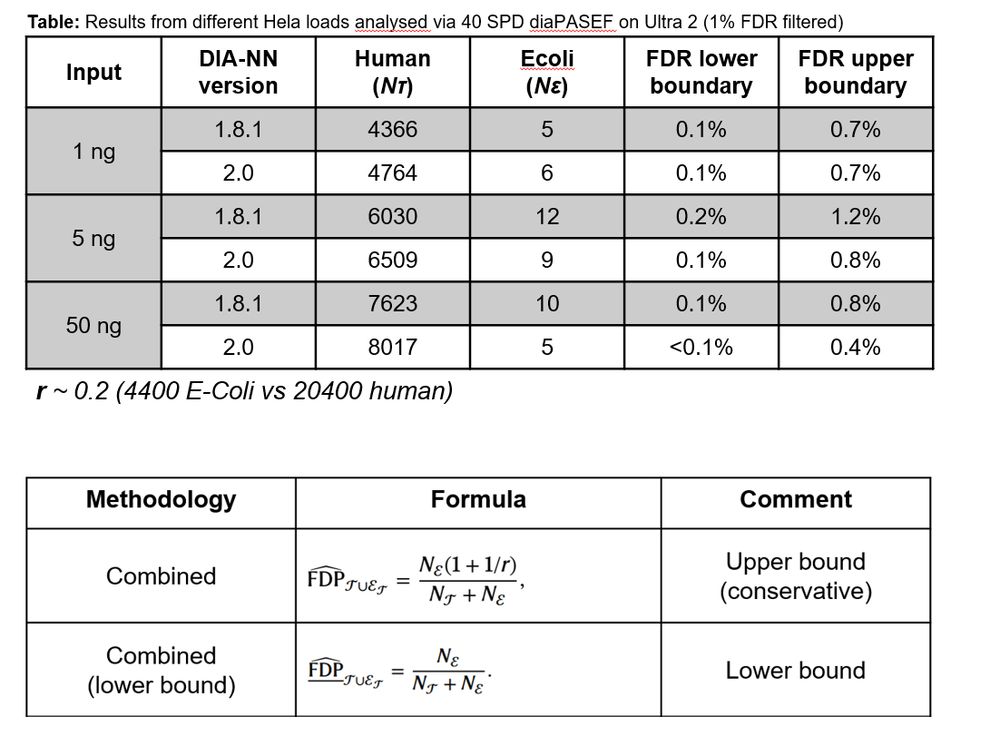




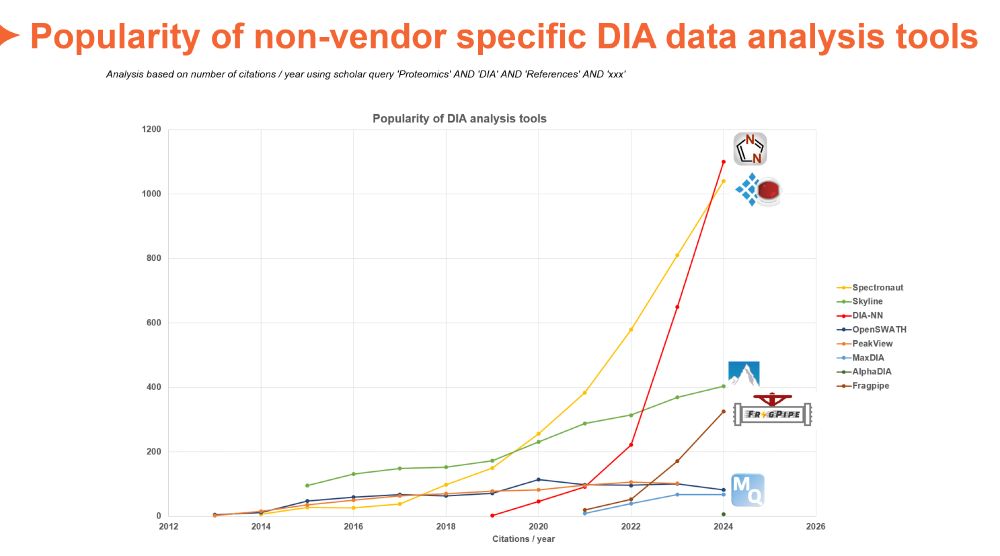
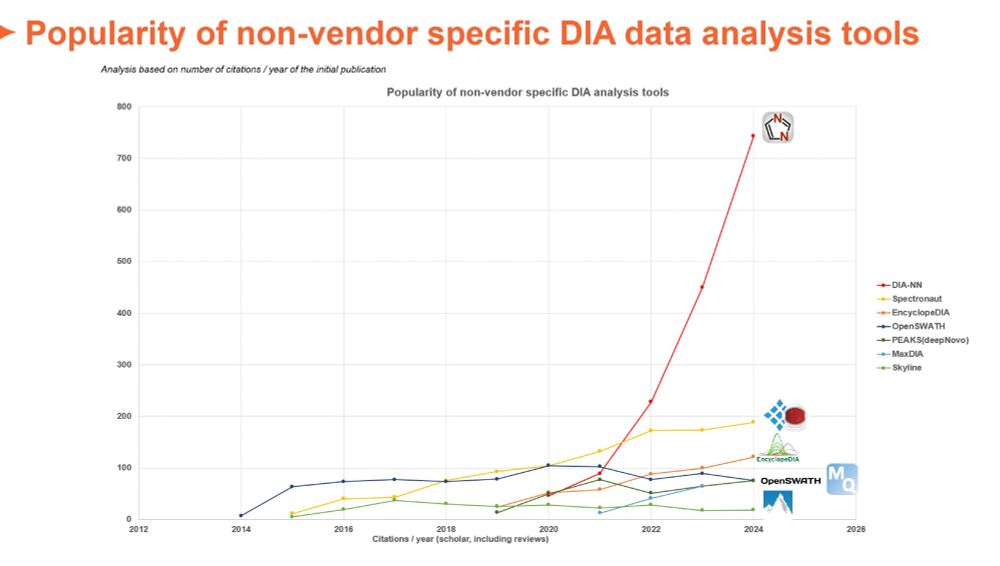

Based on citations/year of initial publication it seems as MSFragger would take over soon...

Based on citations/year of initial publication it seems as MSFragger would take over soon...



