RuthLeyMicro
@microbiome.bsky.social
2.2K followers
200 following
79 posts
Director of Department of Microbiome Science at MPI Biology Tübingen Germany. Also known as Leylab.com.
Posts
Media
Videos
Starter Packs
Pinned
RuthLeyMicro
@microbiome.bsky.social
· Jul 7
Alexander Tyakht
@atyakht.bsky.social
· Jul 7
Human gut flagellome profiling using FlaPro reveals TLR5-related phenotype-specific alterations in IBD
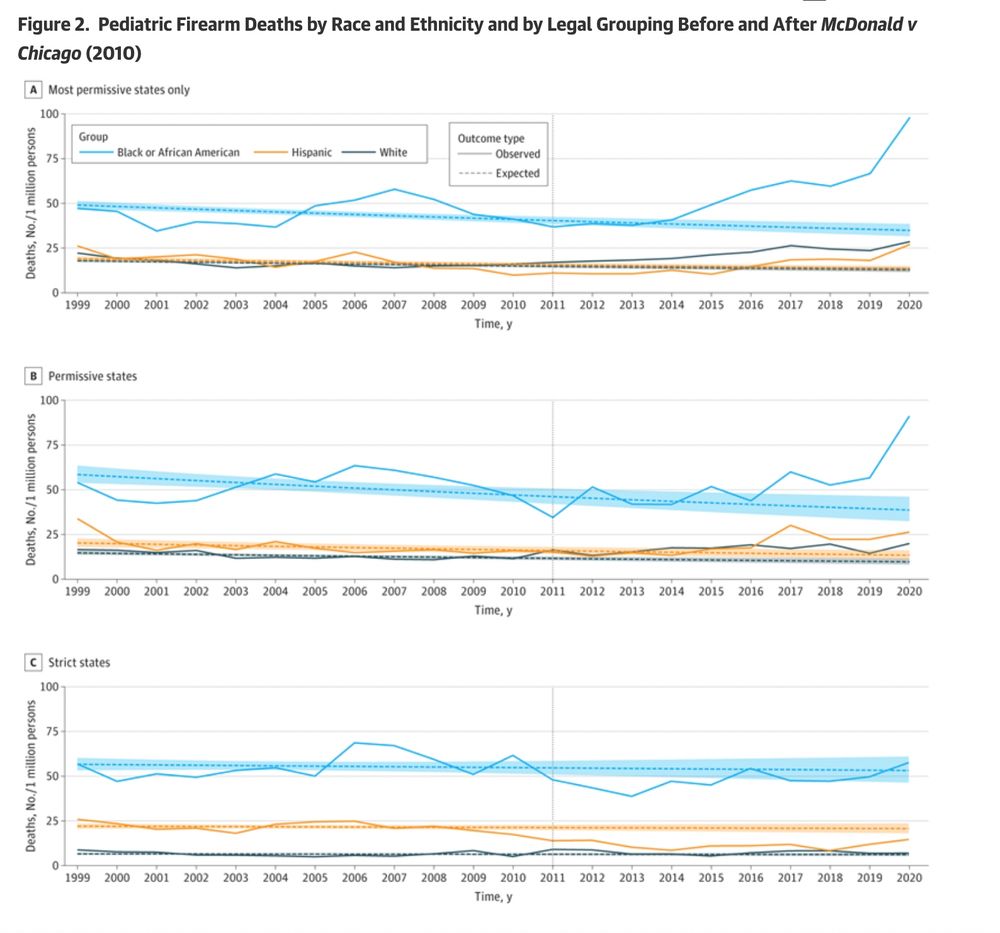
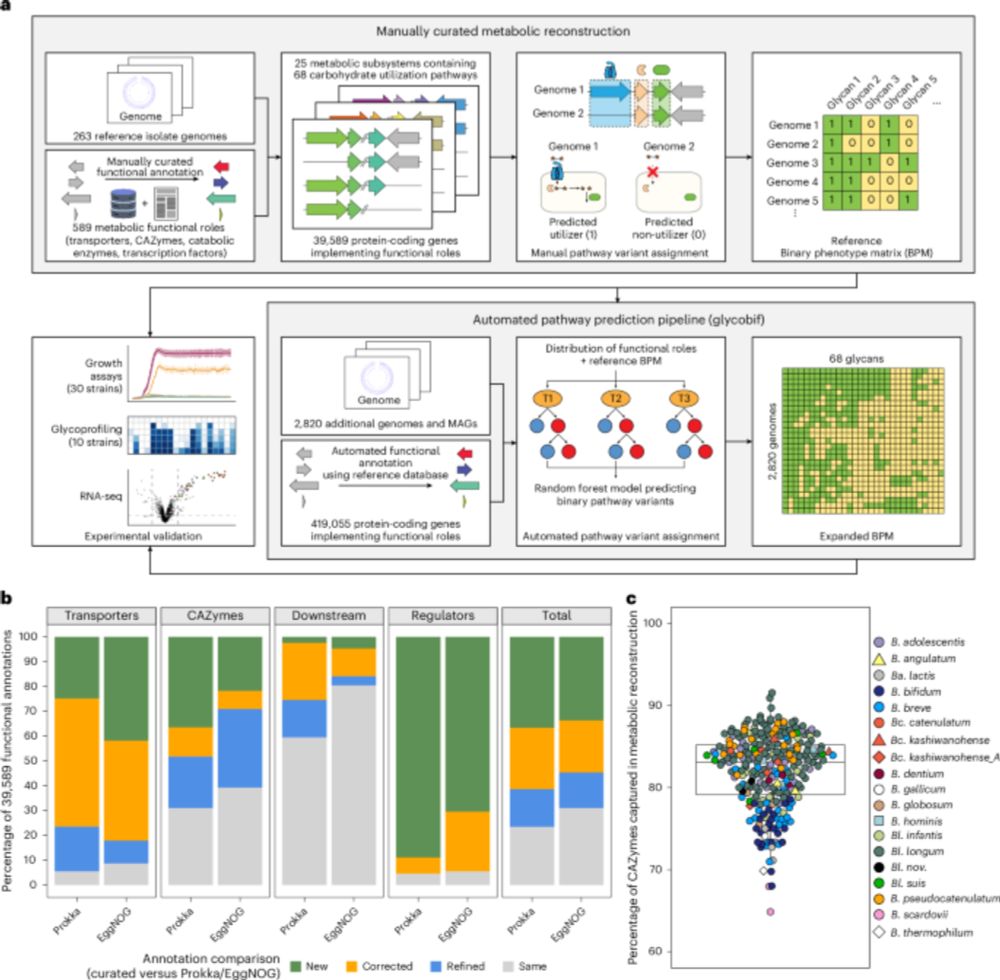
Flagellin is the protein monomer of the bacterial flagellum, which confers motility, allowing bacteria to reach their favored niches. Flagellin is highly conserved across bacterial species and thus the target of the innate immune receptor Toll-like receptor 5 (TLR5). In the gut, bacterial flagellin agonizes human TLR5, triggering a pro-inflammatory response. However, the ability to bind and activate TLR5 varies considerably between different flagellins, suggesting that the composition of an individual's flagellin repertoire - the flagellome - may mediate the inflammatory response to the microbiome, with relevance to inflammatory bowel diseases. However, to date, methods to assess the inflammatory potential of a flagellome are lacking. To address this gap, we constructed a curated database of human gut flagellins. To predict the inflammatory potential of the flagellome by sorting flagellins into either "stimulatory" (strong TLR5 agonists) or "silent" (weak TLR5 agonists), we trained a machine learning model on experimentally characterized flagellins with known binding and stimulatory activities. The FlaPro pipeline was implemented using the Snakemake workflow engine for high-throughput analysis and is available at https://github.com/leylabmpi/FlaPro. To validate our approach and explore clinical associations, we applied FlaPro to a publicly available multi-omics dataset from an inflammatory bowel disease (IBD) cohort. Our analysis demonstrates that FlaPro enables robust profiling of the human gut flagellome from metagenomic and metatranscriptomic data. Analysis of the IBD datasets revealed a depletion of flagellome diversity and a reduced silent-to-stimulatory flagellin abundance ratio in Crohn's disease and ulcerative colitis, observed at both the genomic and transcriptional levels. Multiple condition-specific alterations were identified at the level of individual flagellin clusters. These findings indicate that IBD is associated with distinct alterations in the gut flagellome, particularly in relation to TLR5 recognition. Flagellome features represent a functionally interpretable class of microbiome-derived markers with potential utility in microbiome-wide association studies in the context of human health and disease. ### Competing Interest Statement The authors have declared no competing interest. Max Planck Society and the European Research Council (ERC) under the European Unions Horizon 2020 research and innovation programme Grant agreement
www.biorxiv.org
Reposted by RuthLeyMicro
Reposted by RuthLeyMicro
Reposted by RuthLeyMicro
Reposted by RuthLeyMicro
RuthLeyMicro
@microbiome.bsky.social
· Aug 29
RuthLeyMicro
@microbiome.bsky.social
· Aug 27
Reposted by RuthLeyMicro
Reposted by RuthLeyMicro
Reposted by RuthLeyMicro
RuthLeyMicro
@microbiome.bsky.social
· Jul 30
Reposted by RuthLeyMicro
Reposted by RuthLeyMicro
Reposted by RuthLeyMicro
Matt Olm
@mattolm.bsky.social
· Jul 17
RuthLeyMicro
@microbiome.bsky.social
· Jul 16
Reposted by RuthLeyMicro
RuthLeyMicro
@microbiome.bsky.social
· Jul 7
Alexander Tyakht
@atyakht.bsky.social
· Jul 7
Human gut flagellome profiling using FlaPro reveals TLR5-related phenotype-specific alterations in IBD
Flagellin is the protein monomer of the bacterial flagellum, which confers motility, allowing bacteria to reach their favored niches. Flagellin is highly conserved across bacterial species and thus the target of the innate immune receptor Toll-like receptor 5 (TLR5). In the gut, bacterial flagellin agonizes human TLR5, triggering a pro-inflammatory response. However, the ability to bind and activate TLR5 varies considerably between different flagellins, suggesting that the composition of an individual's flagellin repertoire - the flagellome - may mediate the inflammatory response to the microbiome, with relevance to inflammatory bowel diseases. However, to date, methods to assess the inflammatory potential of a flagellome are lacking. To address this gap, we constructed a curated database of human gut flagellins. To predict the inflammatory potential of the flagellome by sorting flagellins into either "stimulatory" (strong TLR5 agonists) or "silent" (weak TLR5 agonists), we trained a machine learning model on experimentally characterized flagellins with known binding and stimulatory activities. The FlaPro pipeline was implemented using the Snakemake workflow engine for high-throughput analysis and is available at https://github.com/leylabmpi/FlaPro. To validate our approach and explore clinical associations, we applied FlaPro to a publicly available multi-omics dataset from an inflammatory bowel disease (IBD) cohort. Our analysis demonstrates that FlaPro enables robust profiling of the human gut flagellome from metagenomic and metatranscriptomic data. Analysis of the IBD datasets revealed a depletion of flagellome diversity and a reduced silent-to-stimulatory flagellin abundance ratio in Crohn's disease and ulcerative colitis, observed at both the genomic and transcriptional levels. Multiple condition-specific alterations were identified at the level of individual flagellin clusters. These findings indicate that IBD is associated with distinct alterations in the gut flagellome, particularly in relation to TLR5 recognition. Flagellome features represent a functionally interpretable class of microbiome-derived markers with potential utility in microbiome-wide association studies in the context of human health and disease. ### Competing Interest Statement The authors have declared no competing interest. Max Planck Society and the European Research Council (ERC) under the European Unions Horizon 2020 research and innovation programme Grant agreement
www.biorxiv.org
RuthLeyMicro
@microbiome.bsky.social
· Jul 6
RuthLeyMicro
@microbiome.bsky.social
· Jul 5