@miraburtscher.bsky.social
53 followers
51 following
2 posts
Predoc @savitski_lab @EMBL | Computational Biology @saezlab | Interested in functional proteomics and systems biology 🧫🧬
Posts
Media
Videos
Starter Packs
Pinned
Reposted
Reposted
Reposted
Reposted
Savitski Lab
@savitski-lab.bsky.social
· Apr 29

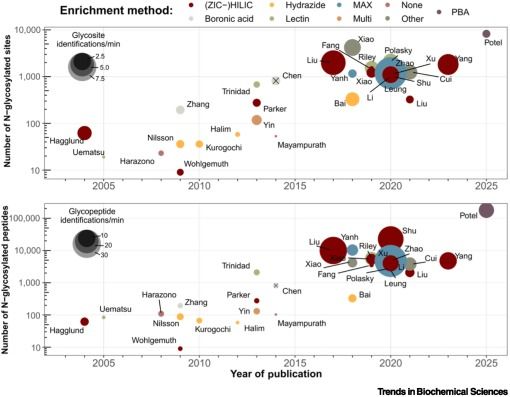
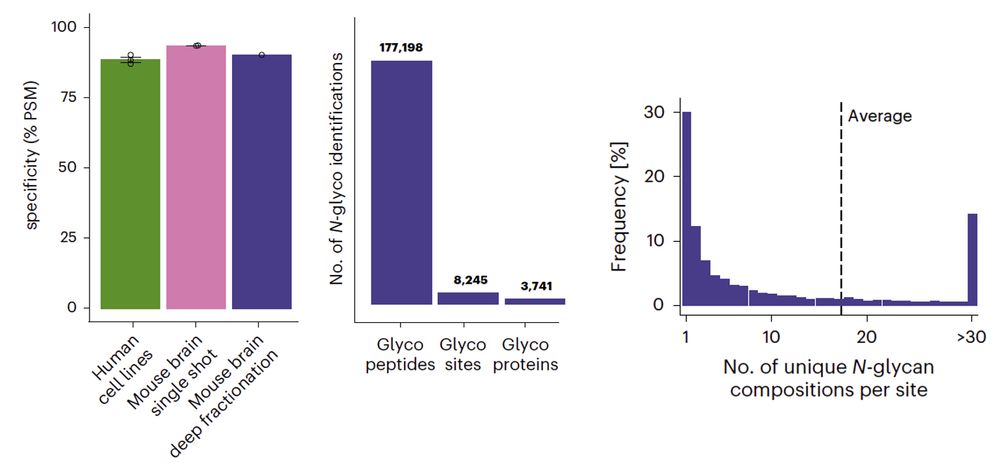
High-throughput peptide-centric local stability assay extends protein-ligand identification to membrane proteins, tissues, and bacteria
Systematic mapping of protein-ligand interactions is essential for understanding biological processes and drug mechanisms. Peptide-centric local stability assay (PELSA) is a powerful tool for detectin...
www.biorxiv.org
Reposted