Savitski Lab
@savitski-lab.bsky.social
310 followers
80 following
17 posts
We are the Savitski lab located @ EMBL using and developing proteomics methods for assessing the state of the proteome
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Savitski Lab
Reposted by Savitski Lab
Reposted by Savitski Lab
Hiroyuki Uechi
@hiro-cg5445.bsky.social
· May 15
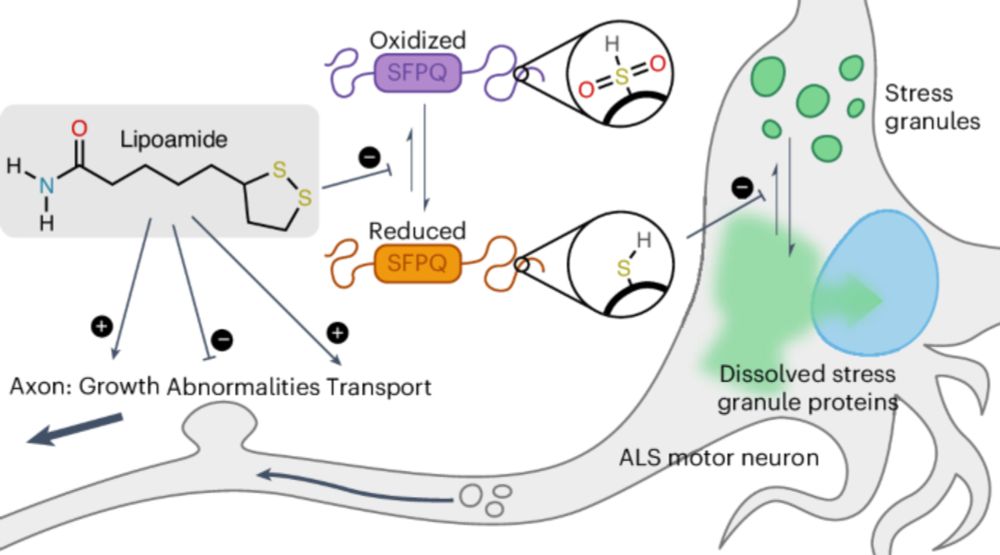
Small-molecule dissolution of stress granules by redox modulation benefits ALS models - Nature Chemical Biology
Uechi et al. found that a small-molecule lipoamide dissolves stress granules (SGs) by targeting SFPQ, a redox-sensitive disordered SG protein, alleviating pathological phenotypes caused by amyotrophic...
www.nature.com
Reposted by Savitski Lab
Hyman Lab
@hymanlab.bsky.social
· May 14
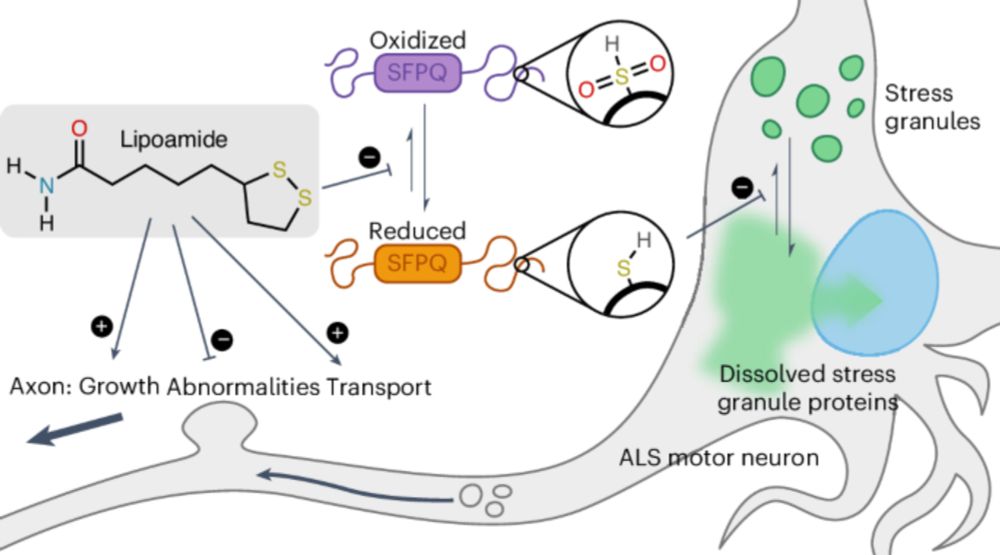
Small-molecule dissolution of stress granules by redox modulation benefits ALS models - Nature Chemical Biology
Uechi et al. found that a small-molecule lipoamide dissolves stress granules (SGs) by targeting SFPQ, a redox-sensitive disordered SG protein, alleviating pathological phenotypes caused by amyotrophic...
www.nature.com
Savitski Lab
@savitski-lab.bsky.social
· Apr 29

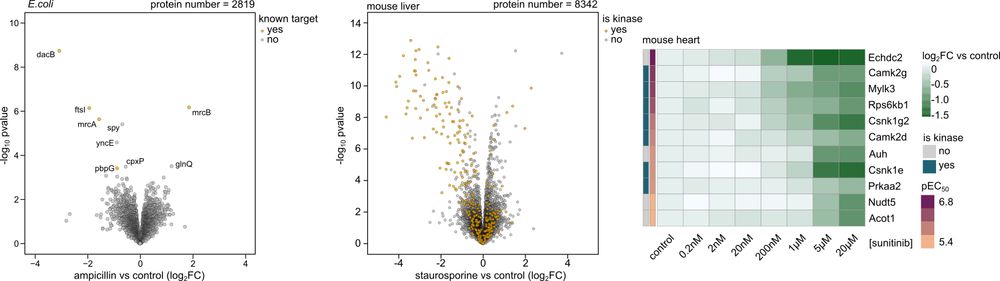
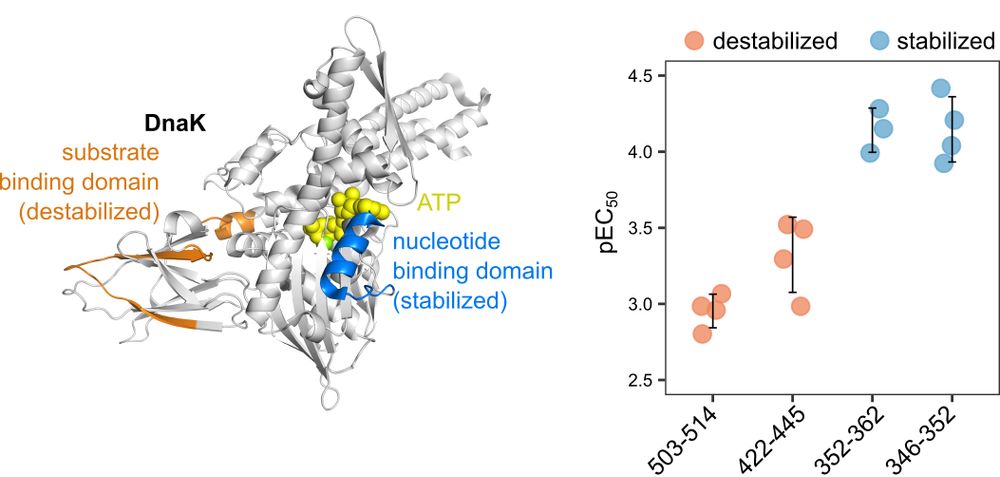
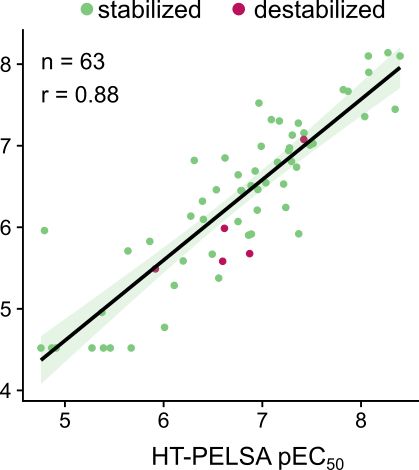
High-throughput peptide-centric local stability assay extends protein-ligand identification to membrane proteins, tissues, and bacteria
Systematic mapping of protein-ligand interactions is essential for understanding biological processes and drug mechanisms. Peptide-centric local stability assay (PELSA) is a powerful tool for detectin...
www.biorxiv.org