• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation
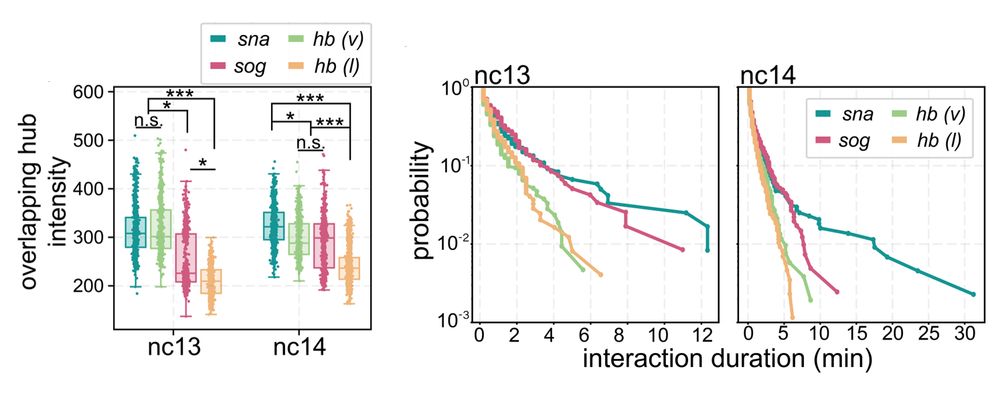
• snail has persistent hub interactions with high-intensity hubs
• sog: has slightly less persistent hub interactions with lower-intensity hubs
• hunchback (negative control): transient interactions without stable hub formation