Nitzan Gonen
@nitzan-gonen.bsky.social
130 followers
120 following
41 posts
Group leader @Bar Ilan University. Interested in sex determination, stem cells and 3D genome organisation. Head of the BIU transgenic unit. ERC StG 2022. Mom of 4.
Posts
Media
Videos
Starter Packs
Nitzan Gonen
@nitzan-gonen.bsky.social
· Jul 27
Nitzan Gonen
@nitzan-gonen.bsky.social
· Jul 27
Nitzan Gonen
@nitzan-gonen.bsky.social
· Jul 27
Nitzan Gonen
@nitzan-gonen.bsky.social
· Jul 27
Nitzan Gonen
@nitzan-gonen.bsky.social
· Jul 27
Nitzan Gonen
@nitzan-gonen.bsky.social
· Jul 27
Nitzan Gonen
@nitzan-gonen.bsky.social
· Jun 27
Reposted by Nitzan Gonen
Daniel Ibrahim
@danielibrahim.bsky.social
· May 27

Conservation of regulatory elements with highly diverged sequences across large evolutionary distances
Nature Genetics - Combining functional genomic data from mouse and chicken with a synteny-based strategy identifies positionally conserved cis-regulatory elements in the absence of direct sequence...
rdcu.be
Reposted by Nitzan Gonen
Evgeny Kvon
@evgenykvon.bsky.social
· Apr 9
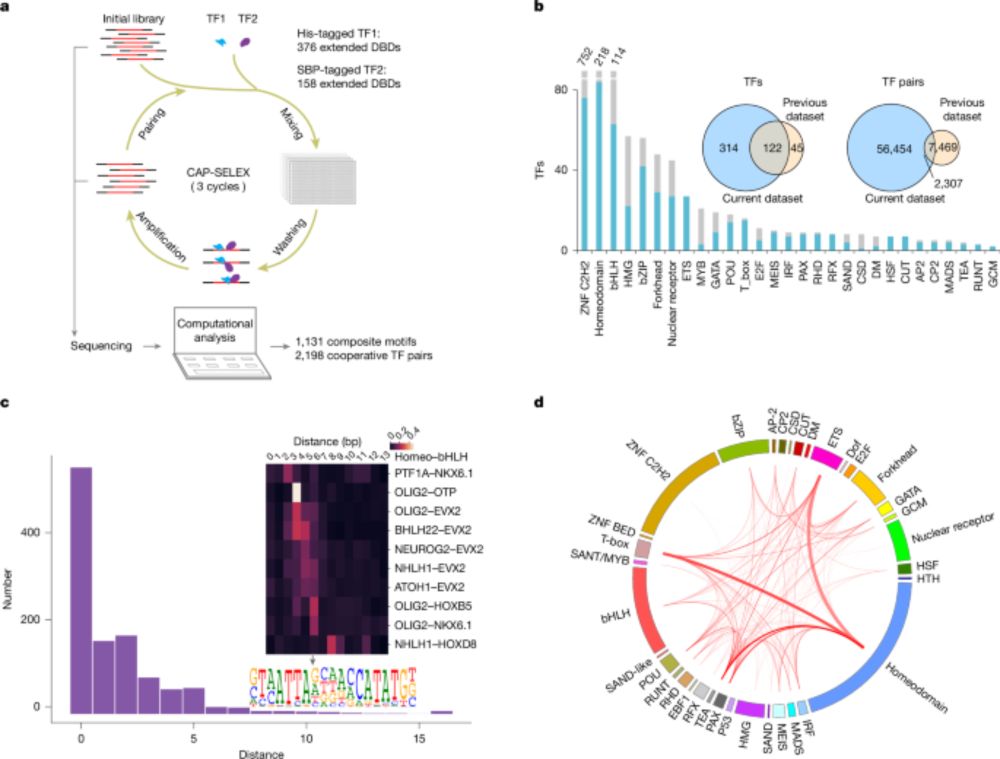
DNA-guided transcription factor interactions extend human gene regulatory code - Nature
A large-scale analysis of DNA-bound transcription factors (TFs) shows how the presence of DNA markedly affects the landscape of TF interactions, and identifies composite motifs that are recognized by ...
www.nature.com