Thomas Norman
@normanlab.bsky.social
82 followers
63 following
16 posts
Assistant Member in csBio at Memorial Sloan Kettering. Perturb-seq, single-cell functional genomics, and techniques for perturbing the genome.
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Thomas Norman
Nature Genetics
@natgenet.nature.com
· Aug 27

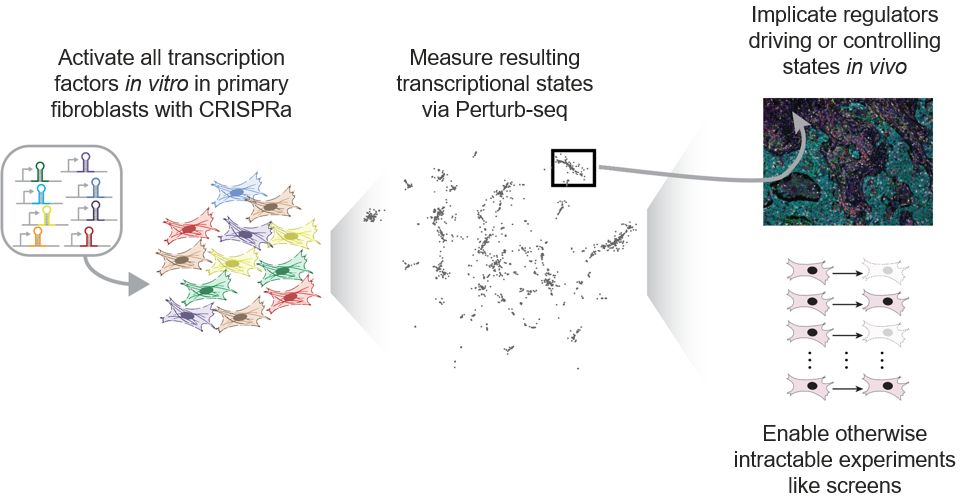
Comprehensive transcription factor perturbations recapitulate fibroblast transcriptional states - Nature Genetics
CRISPR activation of 1,836 human transcription factors recapitulates fibroblast transcriptional states observed in vivo and identifies regulators that can revert inflammatory states.
www.nature.com
Reposted by Thomas Norman
Tommy Vierbuchen
@tvierbuchen.bsky.social
· Aug 27
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6
Thomas Norman
@normanlab.bsky.social
· Aug 6

Comprehensive transcription factor perturbations recapitulate fibroblast transcriptional states
Nature Genetics - CRISPR activation of 1,836 human transcription factors recapitulates fibroblast transcriptional states observed in vivo and identifies regulators that can revert inflammatory states.
rdcu.be