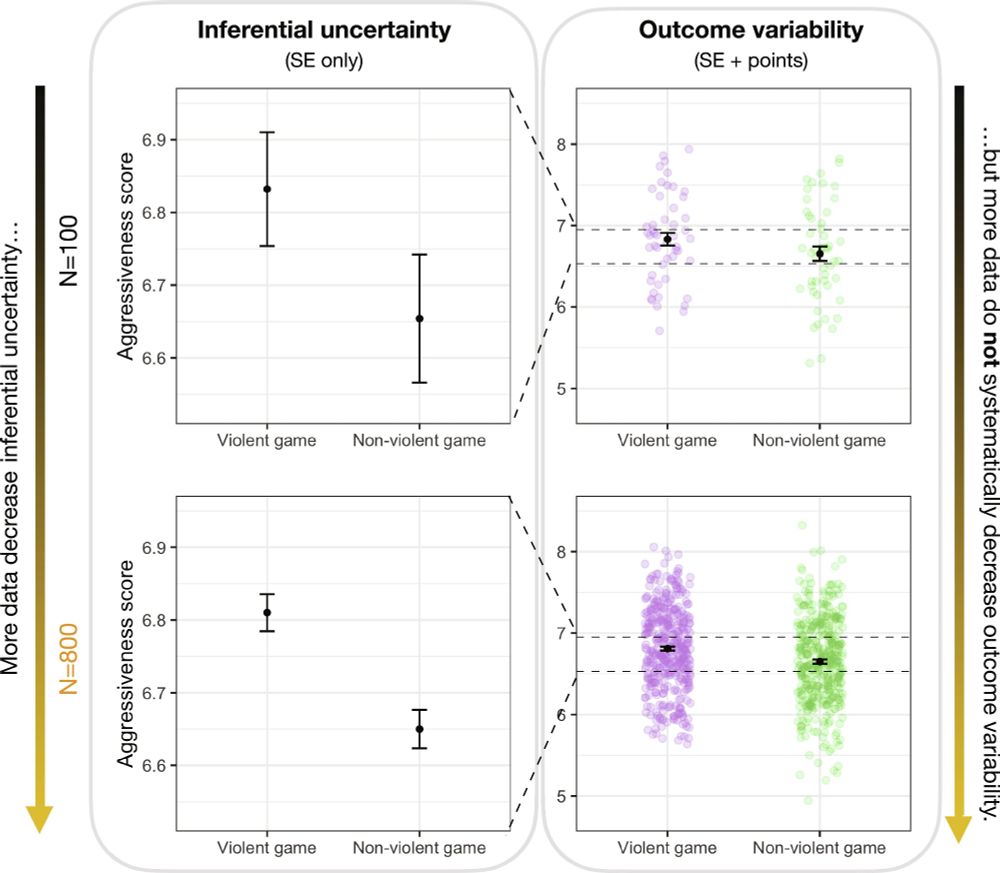
Users save up to 219 hours per year, equivalent to net benefits of up to €5,475 per user.
www.ebi.ac.uk/about/news/a...

Users save up to 219 hours per year, equivalent to net benefits of up to €5,475 per user.
www.ebi.ac.uk/about/news/a...
It enables:
-Label-free multiplexing
-Combinatorial multiplexing with plexDIA
Using combined 9-plexDIA and 3-timePlex we demonstrate 27-plex DIA 🚀
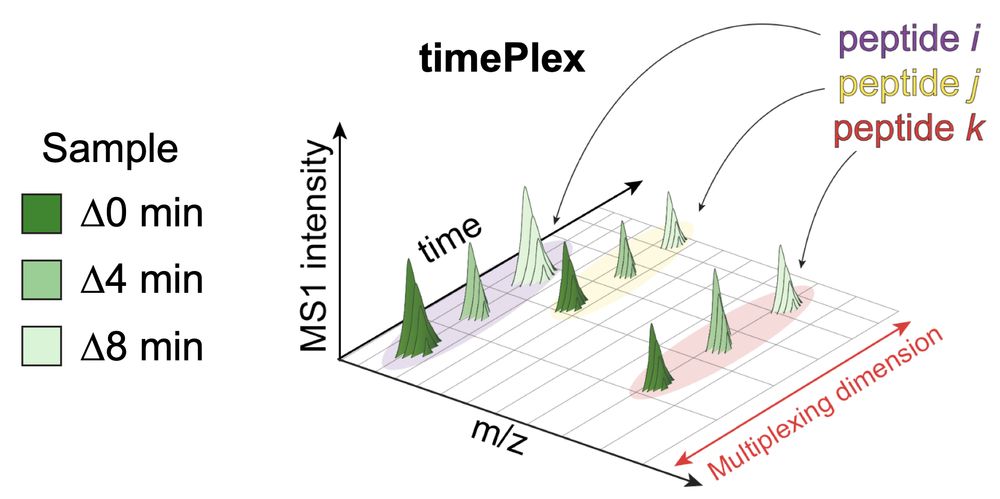
It enables:
-Label-free multiplexing
-Combinatorial multiplexing with plexDIA
Using combined 9-plexDIA and 3-timePlex we demonstrate 27-plex DIA 🚀
We demonstrate 27-plex DIA enabling over 500 samples / day.
We project that timePlex will enable throughputs exceeding 1,000 samples / day.
www.biorxiv.org/content/10.1...
We demonstrate 27-plex DIA enabling over 500 samples / day.
We project that timePlex will enable throughputs exceeding 1,000 samples / day.
www.biorxiv.org/content/10.1...
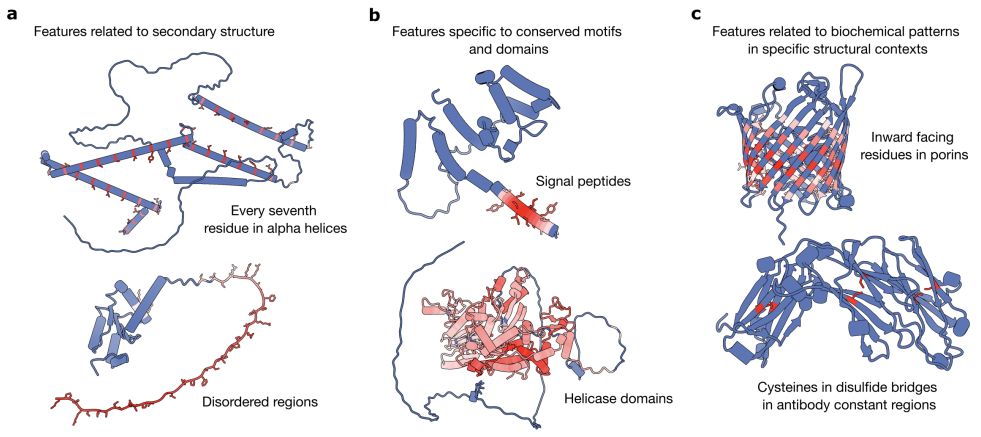
By turning our Jupyter Notebooks into publishable artifacts, we hope to share faster, reduce the effort to convert analysis ➡️ pub, and make our work more reproducible.
Weigh in on our commentary:
research.arcadiascience.com/pub/perspect...

By turning our Jupyter Notebooks into publishable artifacts, we hope to share faster, reduce the effort to convert analysis ➡️ pub, and make our work more reproducible.
Weigh in on our commentary:
research.arcadiascience.com/pub/perspect...
www.nature.com/articles/s41...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Visit web.archive.org
Scroll to 🔎Collection Search section
Select "End Of Term (US Gov 2024)" from the menu
Type in your query term and click search
Or download bulk WARC files from eotarchive.org/data/
#EOT2024 #EOTArchive
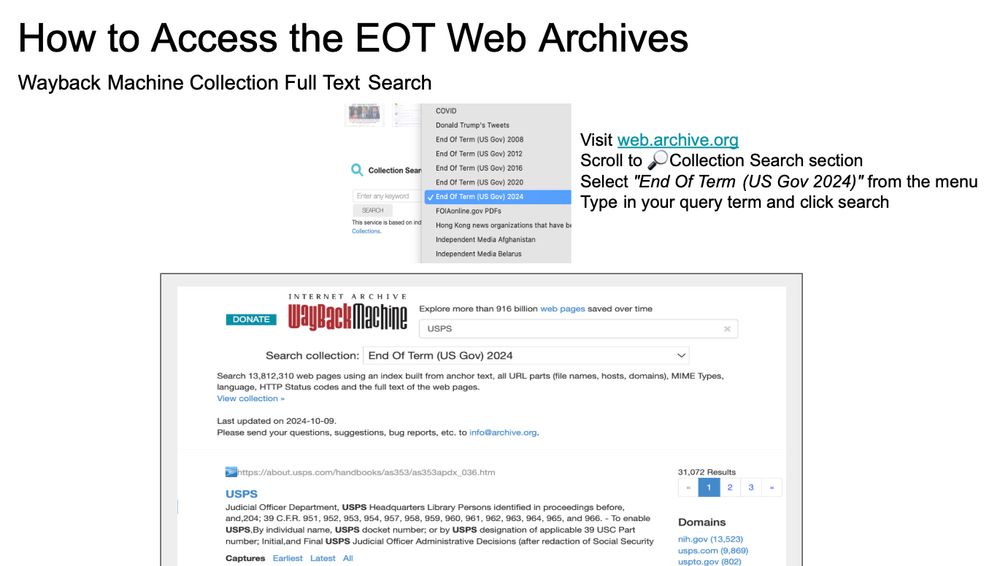
Visit web.archive.org
Scroll to 🔎Collection Search section
Select "End Of Term (US Gov 2024)" from the menu
Type in your query term and click search
Or download bulk WARC files from eotarchive.org/data/
#EOT2024 #EOTArchive
357 validated out of 1,124 tested.
.

357 validated out of 1,124 tested.
.

🧪 🐡 #scicomm

bmj.com/content/331/...
Here's our paper, which shows that experts (doctors, professors, data scientists) mix them up:
www.pnas.org/doi/10.1073/...