Osvaldo Burastero
@osvalb.bsky.social
850 followers
940 following
25 posts
Biophysics enthusiast. Currently focused on data analysis: https://spc.embl-hamburg.de
Posts
Media
Videos
Starter Packs
Reposted by Osvaldo Burastero
Reposted by Osvaldo Burastero
Osvaldo Burastero
@osvalb.bsky.social
· Jun 12
Osvaldo Burastero
@osvalb.bsky.social
· Apr 28
Reposted by Osvaldo Burastero
Clarissa Czekster
@cczekster.bsky.social
· Apr 24

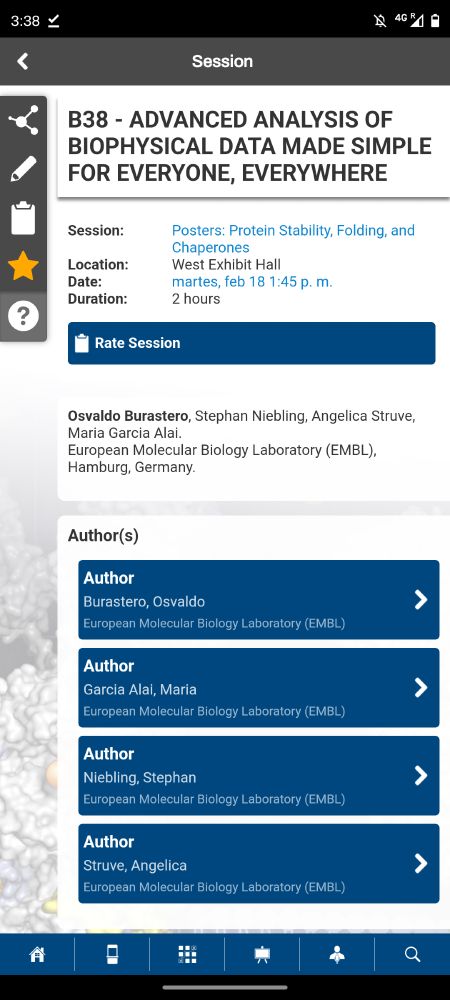
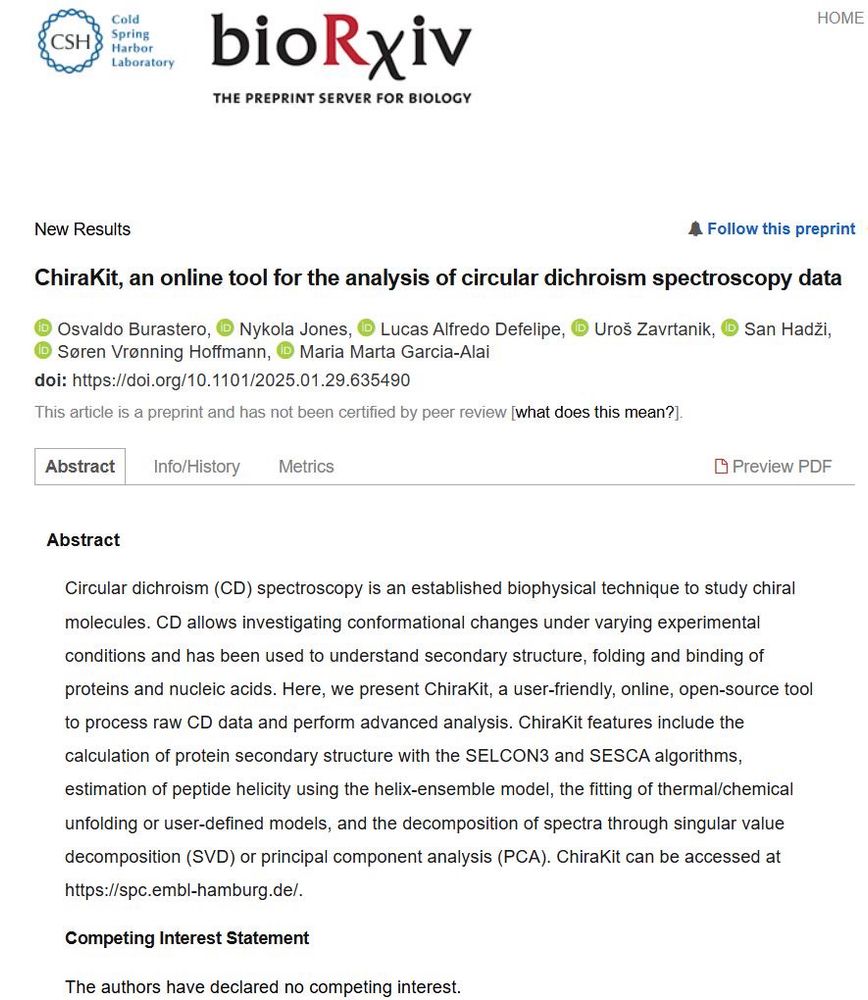
ChiraKit, an online tool for the analysis of circular dichroism spectroscopy data
Circular dichroism (CD) spectroscopy is an established biophysical technique to study chiral molecules. CD allows investigating conformational changes under varying experimental conditions and has bee...
www.biorxiv.org
Osvaldo Burastero
@osvalb.bsky.social
· Jan 31
Osvaldo Burastero
@osvalb.bsky.social
· Jan 31
Reposted by Osvaldo Burastero
Osvaldo Burastero
@osvalb.bsky.social
· Dec 15
Reposted by Osvaldo Burastero
Kate Michie
@kmichie.bsky.social
· Nov 17