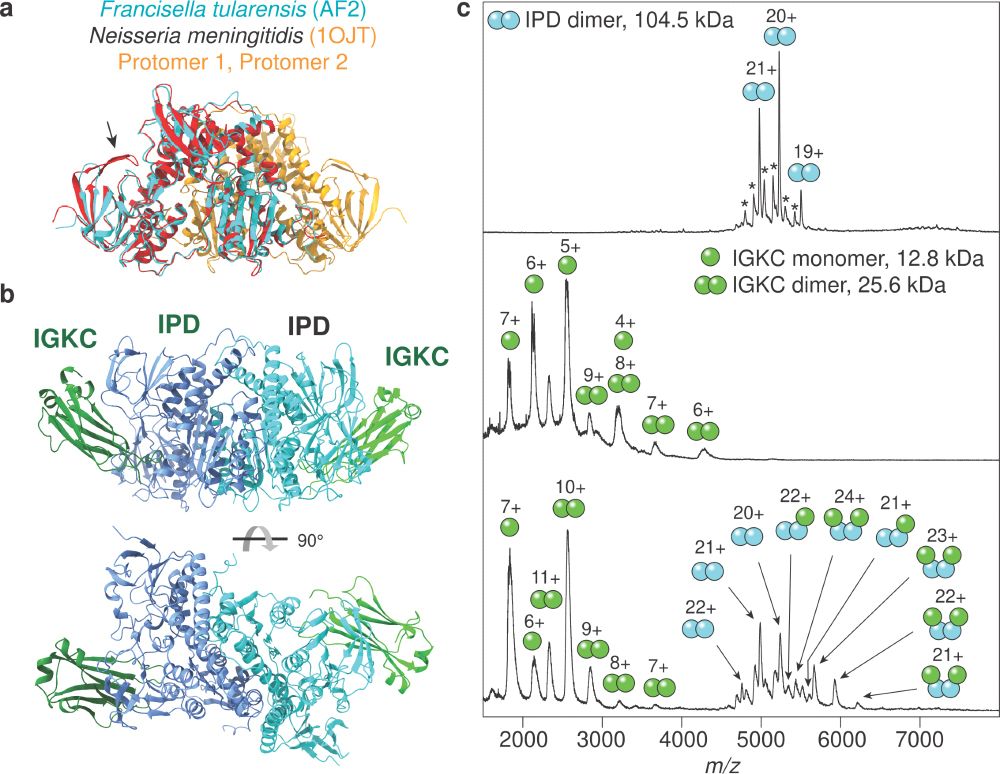
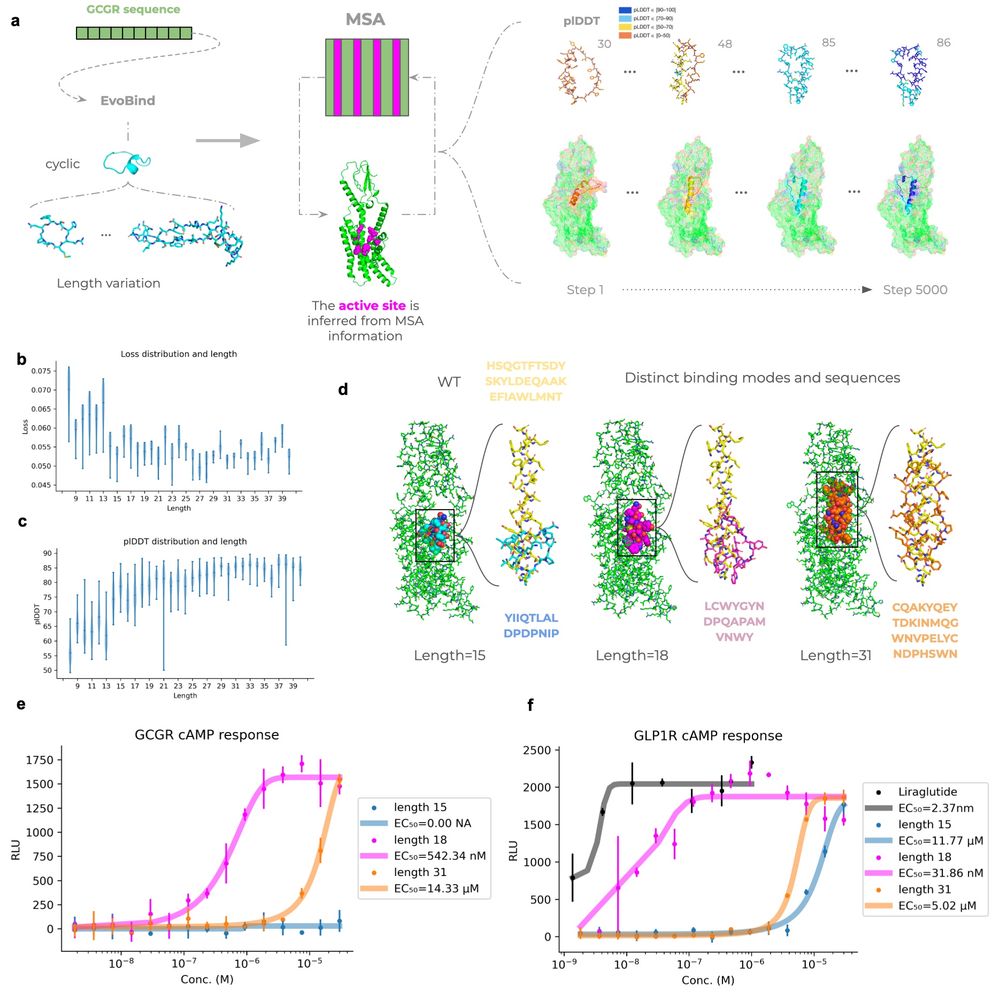
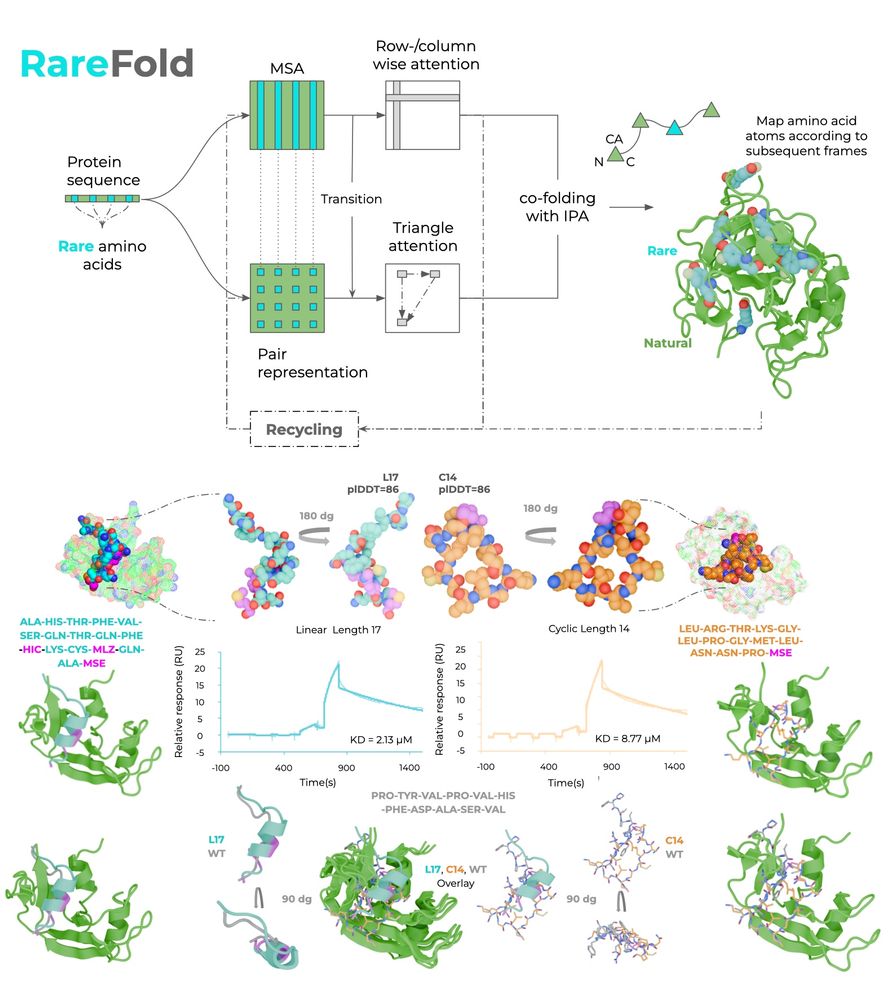
Patrick Bryant
@patrickbryant1.bsky.social
69 followers
26 following
15 posts
Assistant Professor at Stockholm University.
Dedicated Scientist.
Posts
Media
Videos
Starter Packs