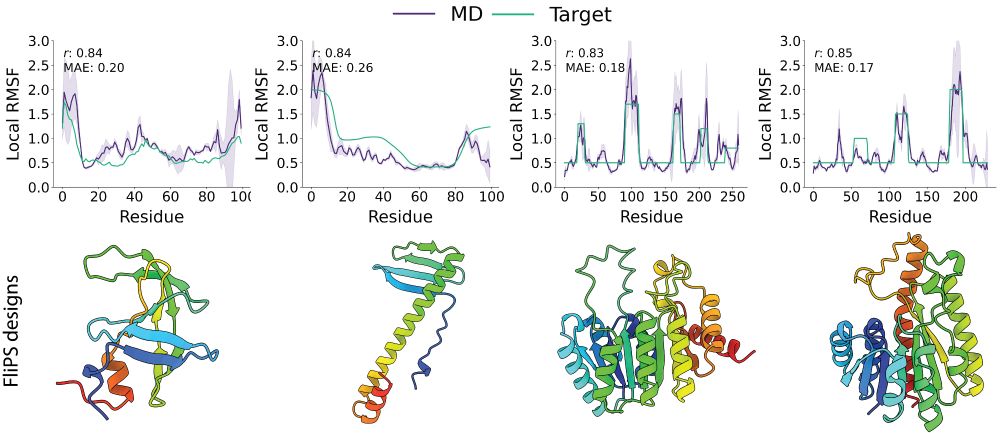
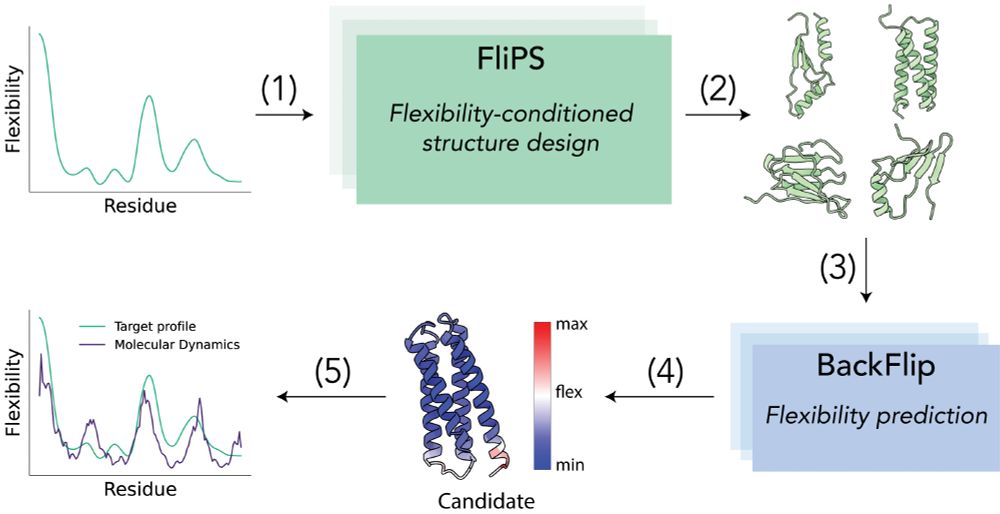
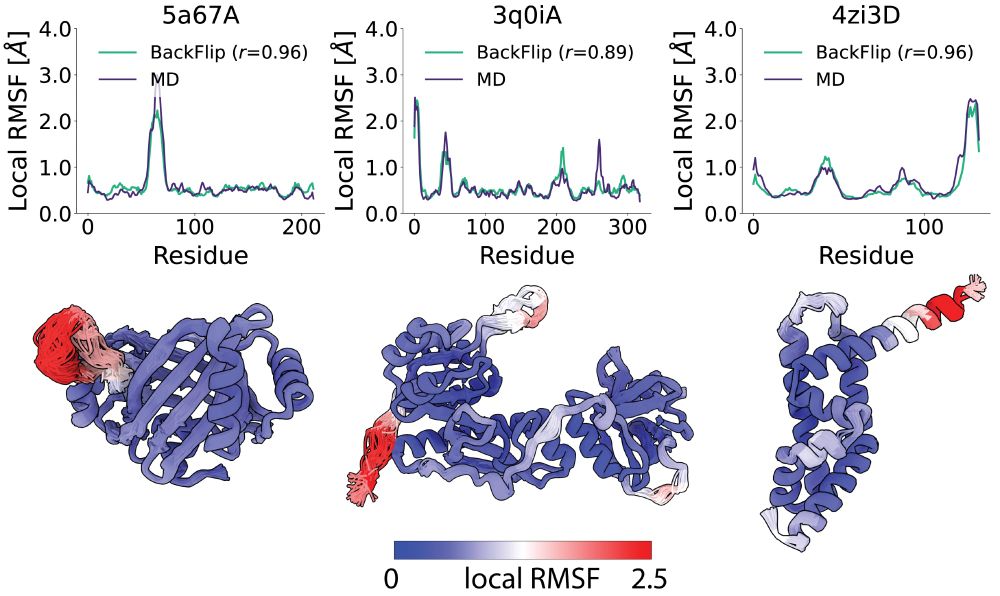
Seva Viliuga
@proteinator.bsky.social
59 followers
96 following
40 posts
PhD candidate in bioinformatics
Protein structure prediction / Protein design
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Seva Viliuga
Seva Viliuga
@proteinator.bsky.social
· Aug 27
Reposted by Seva Viliuga
Reposted by Seva Viliuga
Seva Viliuga
@proteinator.bsky.social
· Jul 17
Seva Viliuga
@proteinator.bsky.social
· Jul 13
Reposted by Seva Viliuga
Seva Viliuga
@proteinator.bsky.social
· Jul 4
Reposted by Seva Viliuga
Seva Viliuga
@proteinator.bsky.social
· Jun 27
Seva Viliuga
@proteinator.bsky.social
· Jun 26
Reposted by Seva Viliuga
Reposted by Seva Viliuga
Reposted by Seva Viliuga
Reposted by Seva Viliuga
Reposted by Seva Viliuga