Paulo C. T. Souza
@pauloctsouza.bsky.social
350 followers
110 following
23 posts
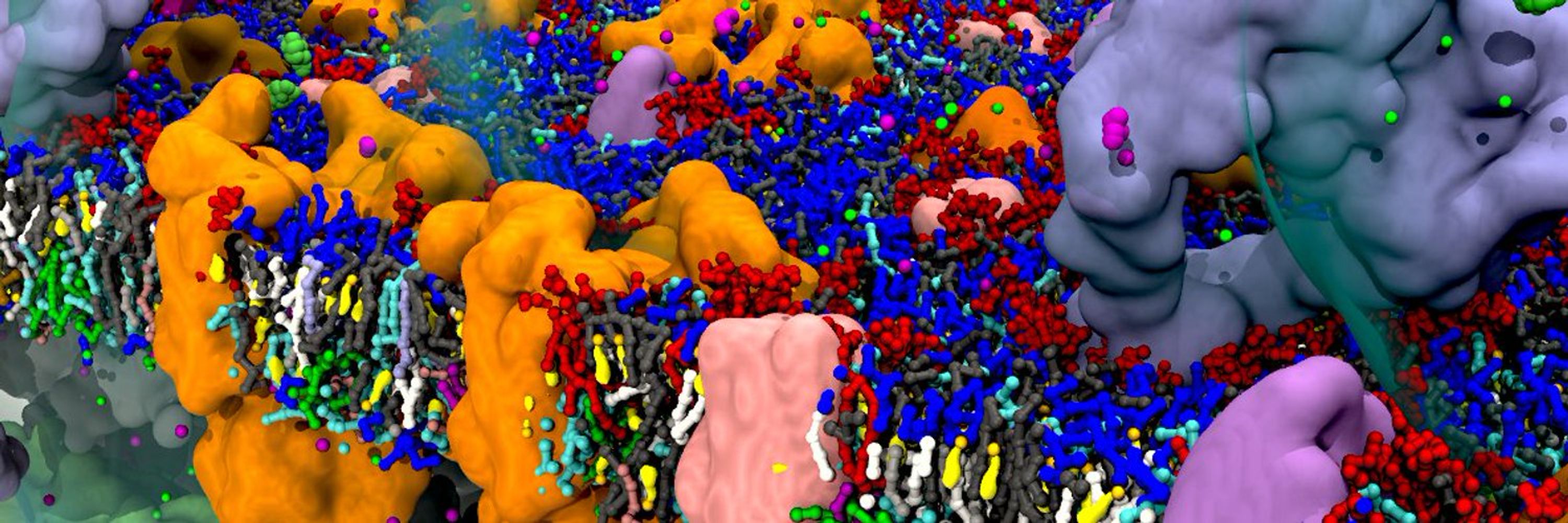
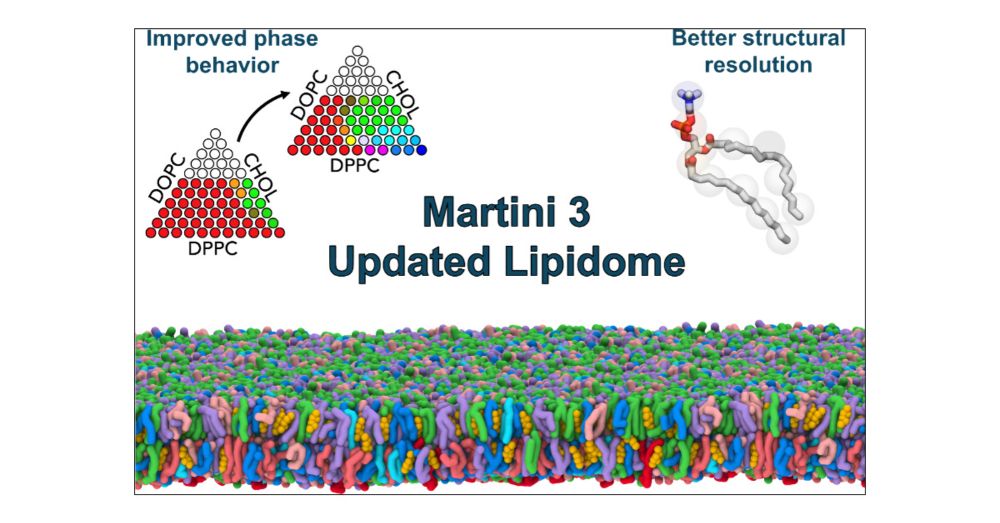
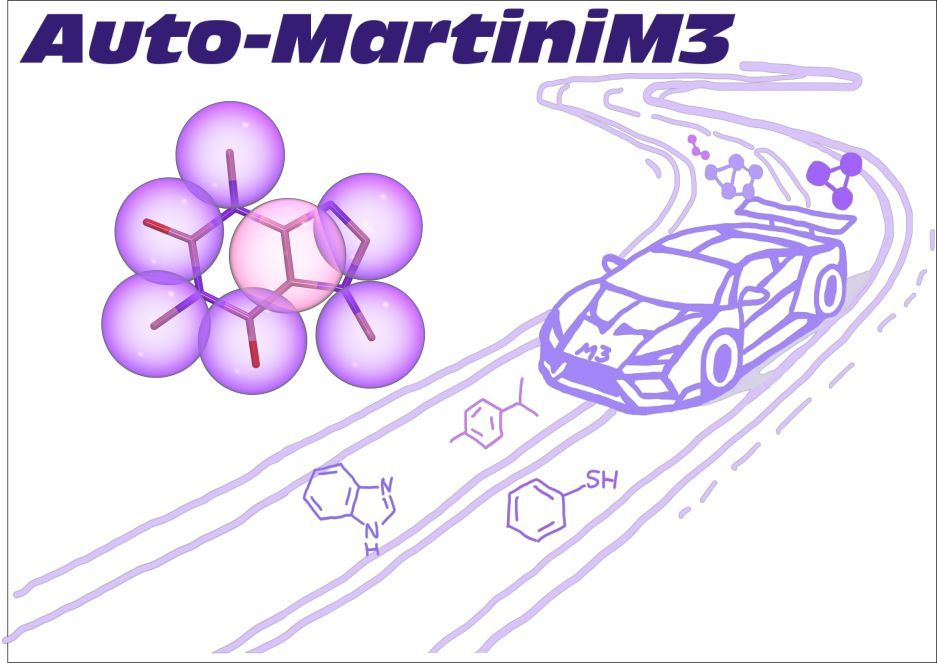
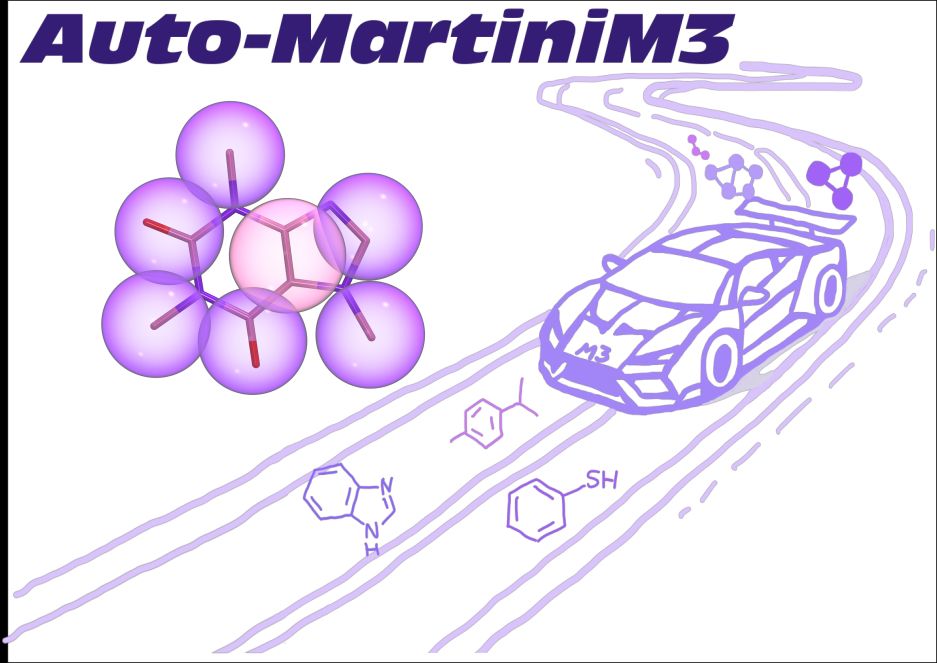
Martini developer trying to make CG models more useful for molecular design and drug/vaccine delivery 🖥️+🍸=💊💉 | Group leader at CBPsmn/ENS de Lyon | CNRS Researcher at DAMM team of LBMC/ENS de Lyon | Scientific consultant
Posts
Media
Videos
Starter Packs
Reposted by Paulo C. T. Souza
Reposted by Paulo C. T. Souza
Reposted by Paulo C. T. Souza
Reposted by Paulo C. T. Souza
Syma Khalid
@sykhalid.bsky.social
· Jan 17
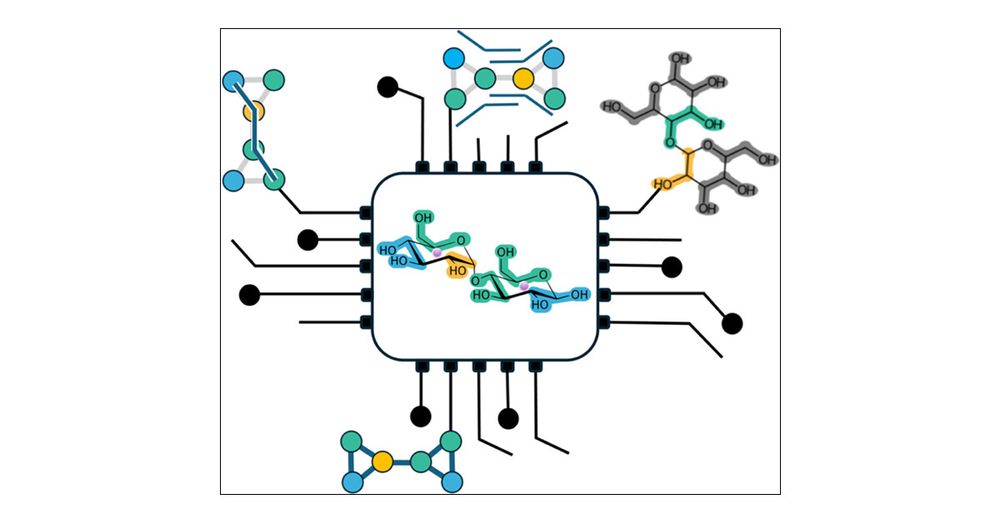
Systematic Approach to Parametrization of Disaccharides for the Martini 3 Coarse-Grained Force Field
Sugars are ubiquitous in biology; they occur in all kingdoms of life. Despite their prevalence, they have often been somewhat neglected in studies of structure–dynamics–function relationships of macro...
pubs.acs.org