Protein structure prediction / Protein design
Preprint: biorxiv.org/content/10.1...
Colab: colab.research.google.com/drive/1KNWvG...
Data: docs.google.com/forms/d/e/1F...

Preprint: biorxiv.org/content/10.1...
Colab: colab.research.google.com/drive/1KNWvG...
Data: docs.google.com/forms/d/e/1F...
@leif-seute.bsky.social, Nicolas Wolf, Simon Wagner, @bioinfo.se, Jan Stühmer and @graeterlab.bsky.social
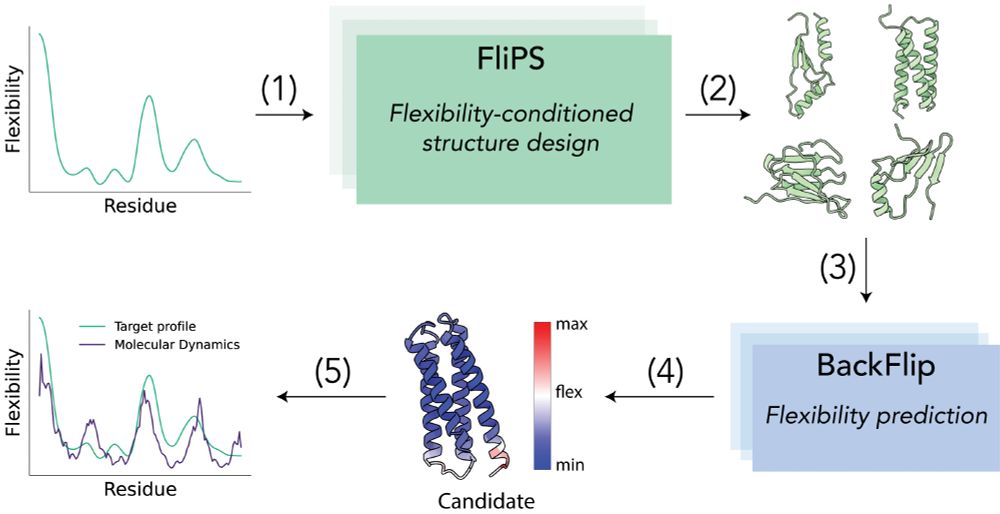
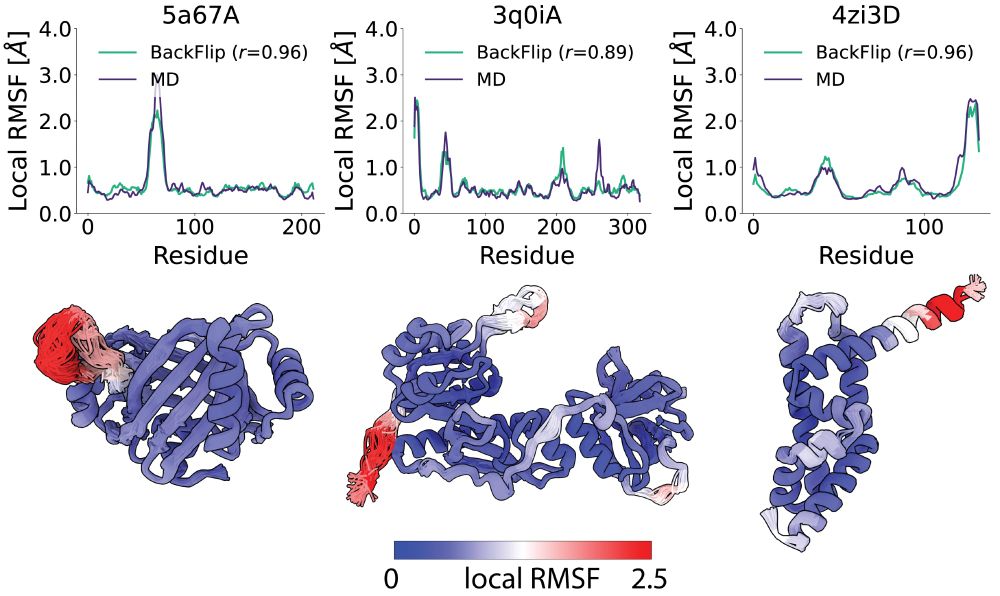
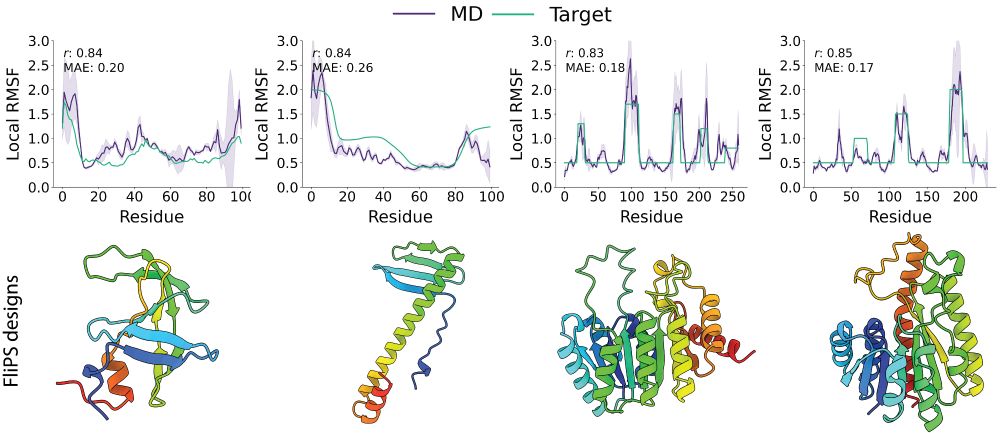
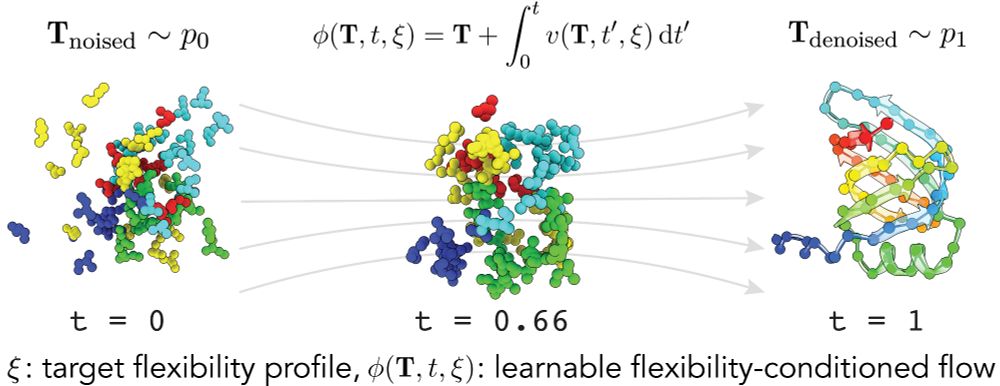
Try out FliPS and BackFlip yourself, code and Google Colab tutorials are available on GitHub!
@leif-seute.bsky.social, Nicolas Wolf, Simon Wagner, @bioinfo.se, Jan Stühmer and @graeterlab.bsky.social
Try out FliPS and BackFlip yourself, code and Google Colab tutorials are available on GitHub!