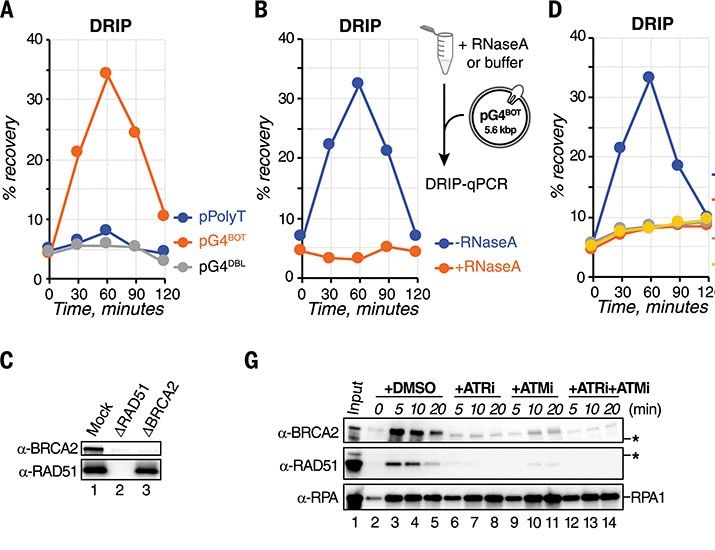
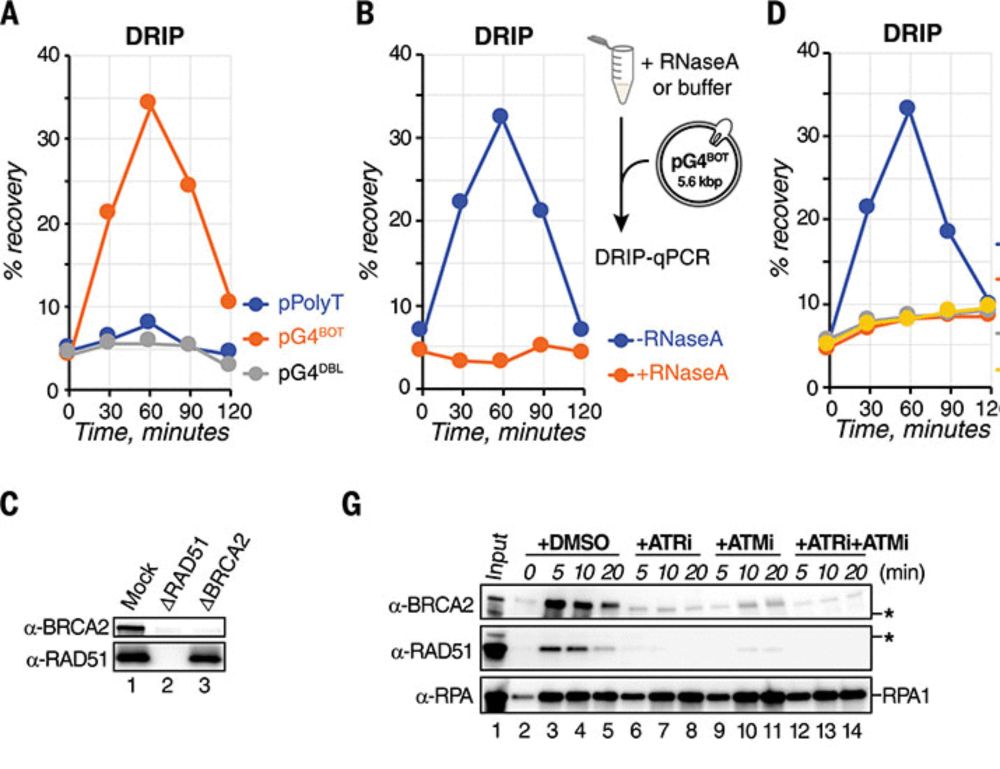
Posts
Media
Videos
Starter Packs
Reposted
Reposted
Reposted
Reposted
Helen Walden
@hw1o9.bsky.social
· Apr 10
Reposted
Reposted