Xuyu Qian
@qianxuyu.bsky.social
120 followers
170 following
19 posts
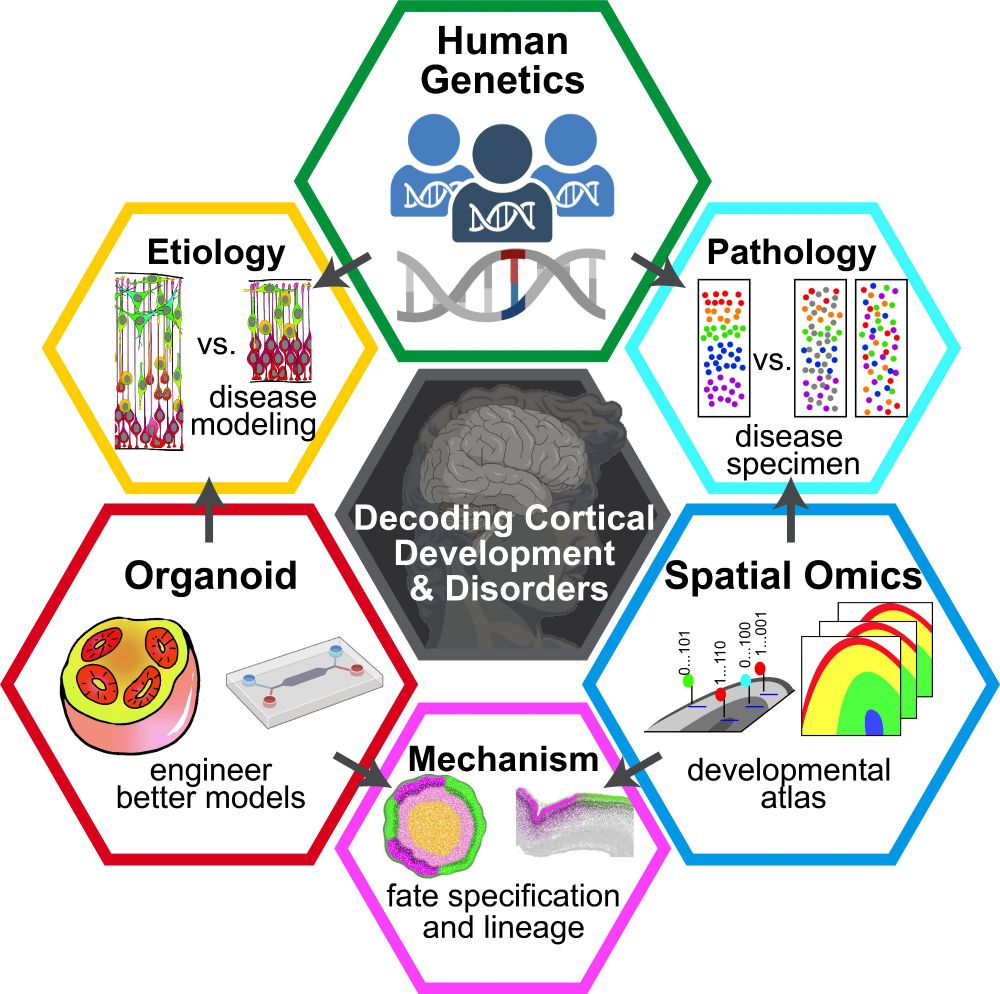
Assist. Prof. @ChildrensPhila & @PennMedicine.
Brain Development , Organoids, Cerebral Cortex.
Postdoc @ChrisAWalsh1; PhD @UPenn_SongMing
el psy congroo.
Posts
Media
Videos
Starter Packs
Pinned
Xuyu Qian
@qianxuyu.bsky.social
· Jul 9
Xuyu Qian
@qianxuyu.bsky.social
· Jul 9

Post-Doctoral Research Fellow - Neurology in Philadelphia, Pennsylvania, United States of America | Fellows, Students & Interns at Children’s Hospital of Philadelphia
Apply for Post-Doctoral Research Fellow - Neurology job with Children’s Hospital of Philadelphia in Philadelphia, Pennsylvania, United States of America. Fellows, Students & Interns at Children’s Hosp...
careers.chop.edu
Xuyu Qian
@qianxuyu.bsky.social
· Jun 2
Xuyu Qian
@qianxuyu.bsky.social
· May 28
Xuyu Qian
@qianxuyu.bsky.social
· May 19
Xuyu Qian
@qianxuyu.bsky.social
· May 14
Xuyu Qian
@qianxuyu.bsky.social
· May 14
Xuyu Qian
@qianxuyu.bsky.social
· May 14
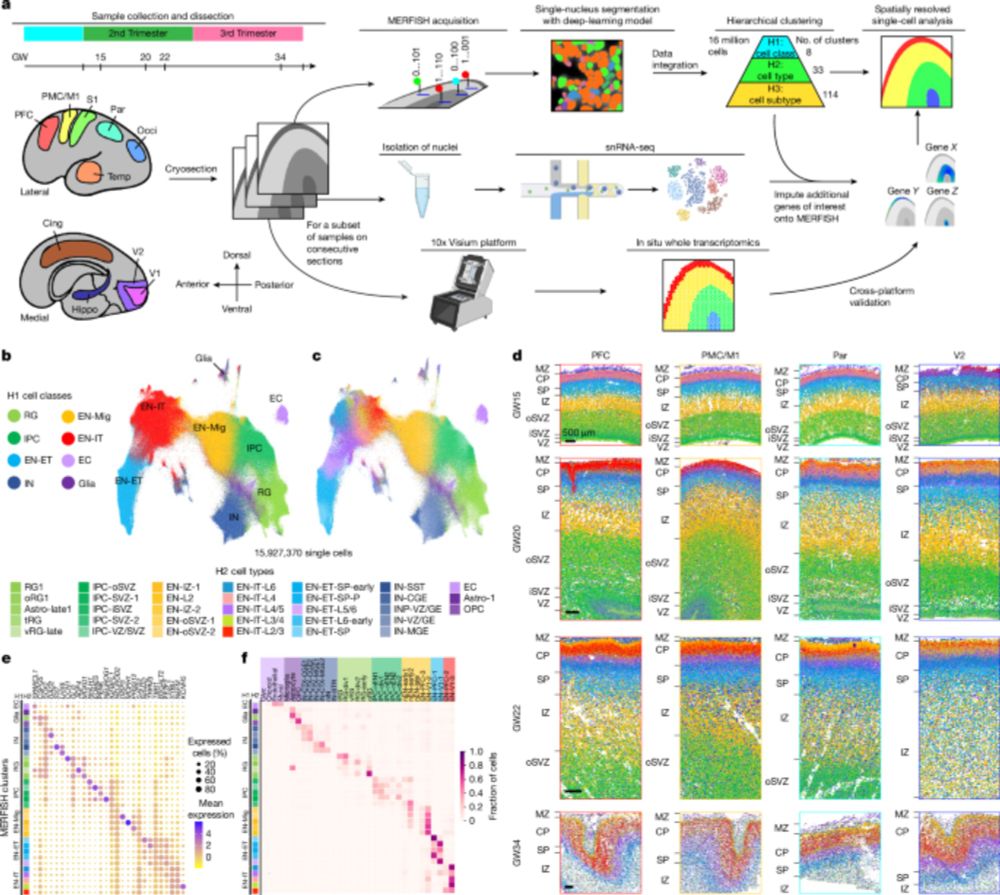
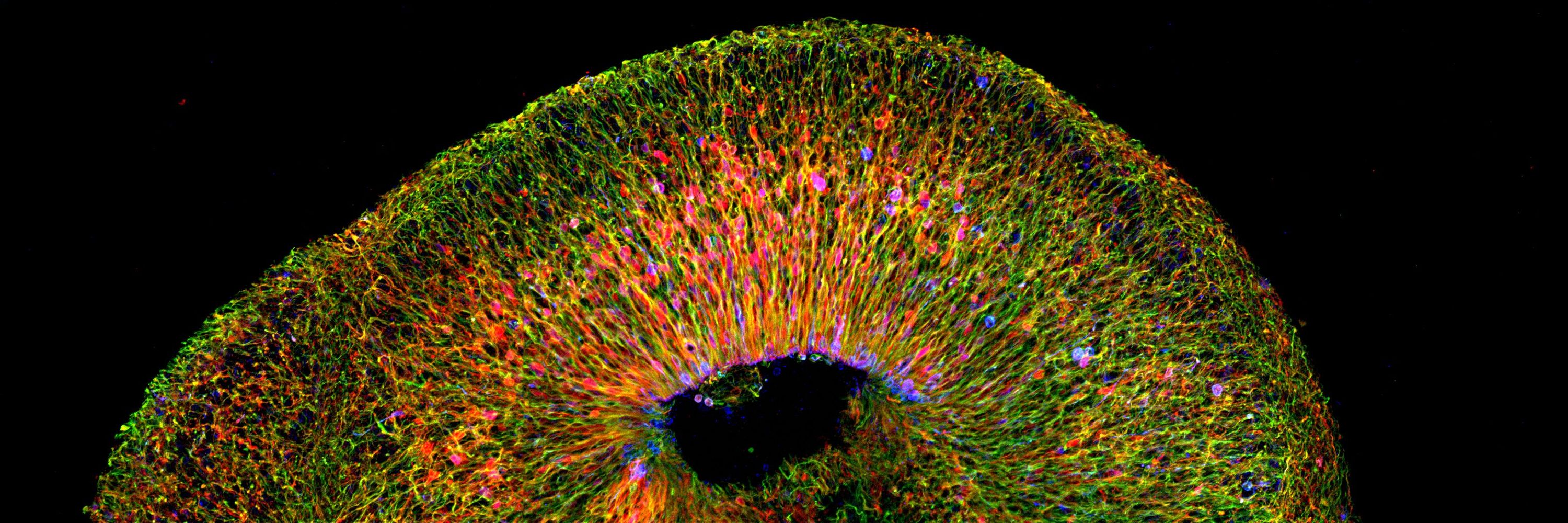
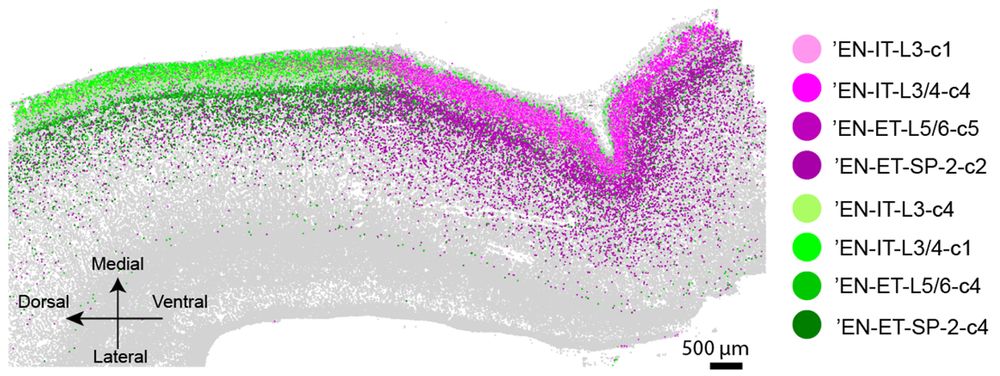
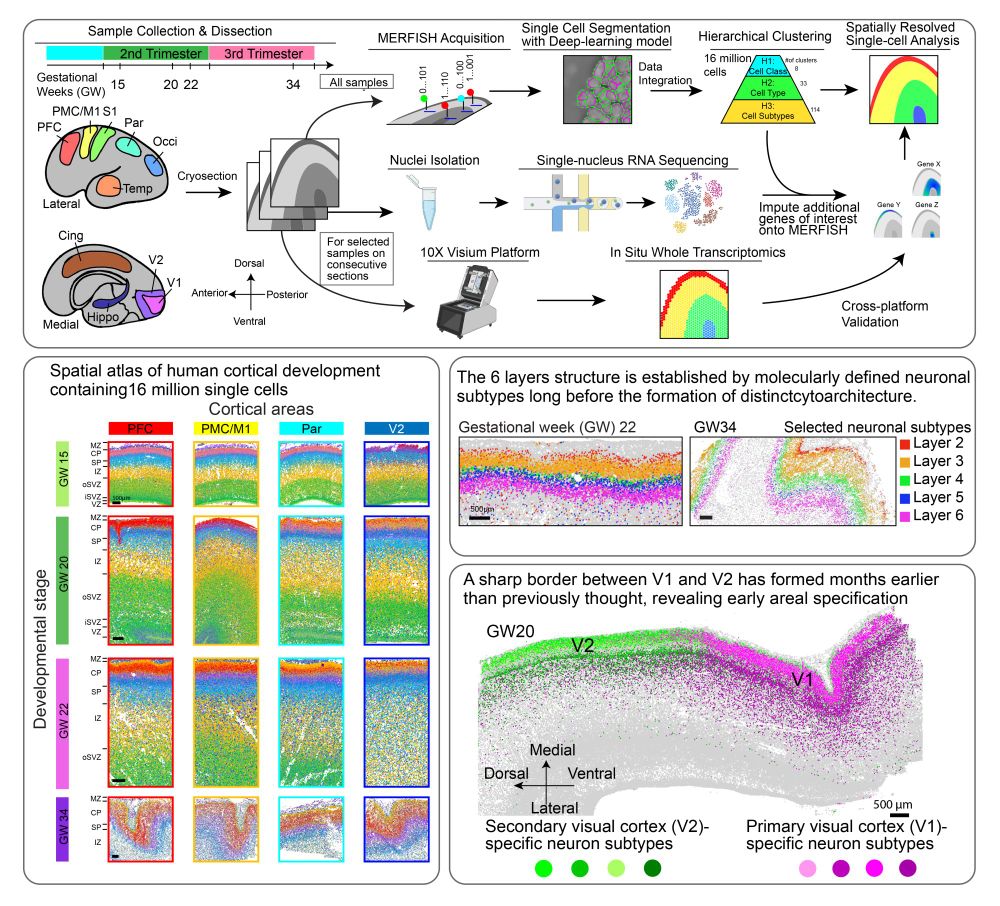
Spatial transcriptomics reveals human cortical layer and area specification - Nature
Multiplexed error-robust fluorescence in situ hybridization (MERFISH) together with deep-learning-based nucleus segmentation enabled the construction of a highly detailed and informative spatially res...
www.nature.com
Reposted by Xuyu Qian
Janet Song
@janetsong.bsky.social
· Apr 2

Human-chimpanzee tetraploid system defines mechanisms of species-specific neural gene regulation
A major challenge in human evolutionary biology is to pinpoint genetic differences that underlie human-specific traits, such as increased neuron number and differences in cognitive behaviors. We used human-chimpanzee tetraploid cells to distinguish gene expression changes due to cis -acting sequence variants that change local gene regulation, from trans expression changes due to species differences in the cellular environment. In neural progenitor cells, examination of both cis and trans changes – combined with CRISPR inhibition and transcription factor motif analyses – identified cis -acting, species-specific gene regulatory changes, including to TNIK , FOSL2 , and MAZ , with widespread trans effects on neurogenesis-related gene programs. In excitatory neurons, we identified POU3F2 as a key cis -regulated gene with trans effects on synaptic gene expression and neuronal firing. This study identifies cis -acting genomic changes that cause cascading trans gene regulatory effects to contribute to human neural specializations, and provides a general framework for discovering genetic differences underlying human traits. ### Competing Interest Statement C.A.W. is on the SAB of Bioskyrb Genomics (cash, equity) and Mosaica Therapeutics (cash, equity), and is an advisor to Maze Therapeutics (equity), but these have no relevance to this work. The remaining authors declare no competing interests.
www.biorxiv.org
Reposted by Xuyu Qian
Reposted by Xuyu Qian
Zinan Zhou
@zinan-zhou.bsky.social
· Mar 13

Recurrent patterns of widespread neuronal genomic damage shared by major neurodegenerative disorders
Amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and Alzheimer's disease (AD) are common neurodegenerative disorders for which the mechanisms driving neuronal death remain unclear. ...
doi.org
Reposted by Xuyu Qian