See example:
biorxiv.org/content/10.1...
Zhidian Zhang @yoakiyama.bsky.social @yehlincho.bsky.social @jajoosam.bsky.social
(3/4)

See example:
biorxiv.org/content/10.1...
Zhidian Zhang @yoakiyama.bsky.social @yehlincho.bsky.social @jajoosam.bsky.social
(3/4)
py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)
Introducing: py2Dmol 🧬
(feedback, suggestions, requests are welcome)

Introducing: py2Dmol 🧬
(feedback, suggestions, requests are welcome)




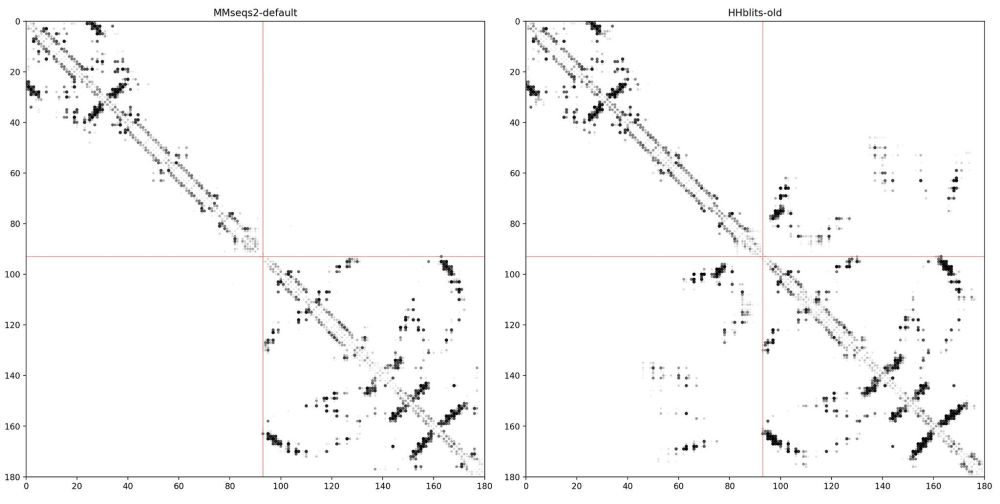
@yoakiyama.bsky.social and Zhidian Zhang on MSA pairformer. (1/4)
@yoakiyama.bsky.social and Zhidian Zhang on MSA pairformer. (1/4)

www.cell.com/cell/abstrac...

www.cell.com/cell/abstrac...