Teun Huijben
@teunhuijben.bsky.social
1.8K followers
490 following
55 posts
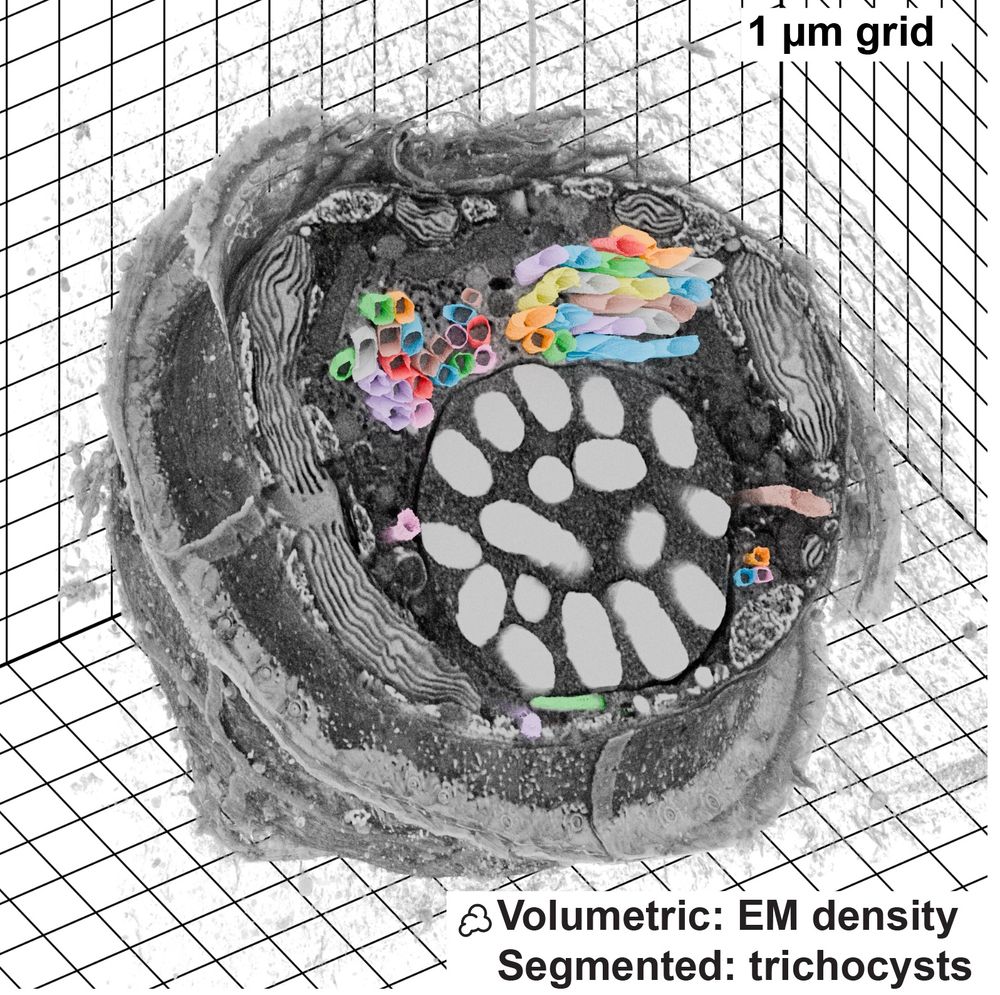
Scientist/PostDoc with Loïc Royer at Chan Zuckerberg Biohub, San Francisco 🔬 image analysis | localization microscopy | machine learning, 🇳🇱 by bike 🚲
📍San Francisco
Posts
Media
Videos
Starter Packs
Pinned
Teun Huijben
@teunhuijben.bsky.social
· Nov 12
Reposted by Teun Huijben
Reposted by Teun Huijben
Teun Huijben
@teunhuijben.bsky.social
· Dec 11
Teun Huijben
@teunhuijben.bsky.social
· Dec 5
Reposted by Teun Huijben