Tal Korem
@tkorem.bsky.social
490 followers
420 following
77 posts
Microbiome, metagenomics, ML, and reproductive health. All views are mine. So are all your base
Posts
Media
Videos
Starter Packs
Pinned
Tal Korem
@tkorem.bsky.social
· Aug 7
Korem Lab - Microbiome Systems Biology @ Columbia | New York, USA
We apply systems biology approaches to decipher the metabolic interactions between the microbiome and its human host in diverse clinical settings, aiming towards personalized microbiome-based therapeu...
www.koremlab.science
Tal Korem
@tkorem.bsky.social
· 19d
Tal Korem
@tkorem.bsky.social
· 19d
Tal Korem
@tkorem.bsky.social
· Aug 16
Annalee
@flowerhorne.com
· Aug 15
Tal Korem
@tkorem.bsky.social
· Aug 7

Korem Lab - Microbiome Systems Biology @ Columbia | New York, USA
We apply systems biology approaches to decipher the metabolic interactions between the microbiome and its human host in diverse clinical settings, aiming towards personalized microbiome-based therapeu...
www.koremlab.science
Reposted by Tal Korem
Tal Korem
@tkorem.bsky.social
· Jul 17
Reposted by Tal Korem
Noah Fierer
@noahfierer.bsky.social
· Jun 21
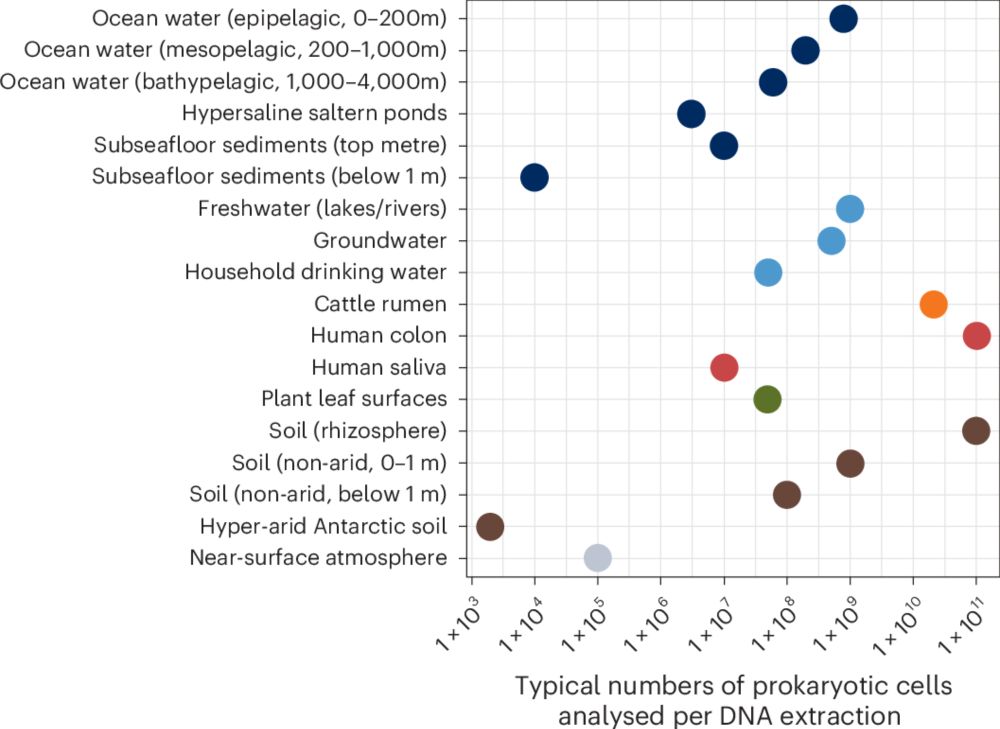
Guidelines for preventing and reporting contamination in low-biomass microbiome studies
Nature Microbiology - In this Consensus Statement, the authors outline strategies for processing, analysing and interpreting low-biomass microbiome samples, and provide recommendations to minimize...
rdcu.be
Reposted by Tal Korem
Reposted by Tal Korem
Reposted by Tal Korem
"spooky sukkot" daniel s.
@self.agency
· May 18
Evan Torner
@guyintheblackhat.bsky.social
· May 18
Reposted by Tal Korem
Tal Korem
@tkorem.bsky.social
· May 2
Tal Korem
@tkorem.bsky.social
· May 2
Reposted by Tal Korem
Emily White
@emily-white.bsky.social
· Apr 11
Tal Korem
@tkorem.bsky.social
· Apr 4
Tal Korem
@tkorem.bsky.social
· Mar 27
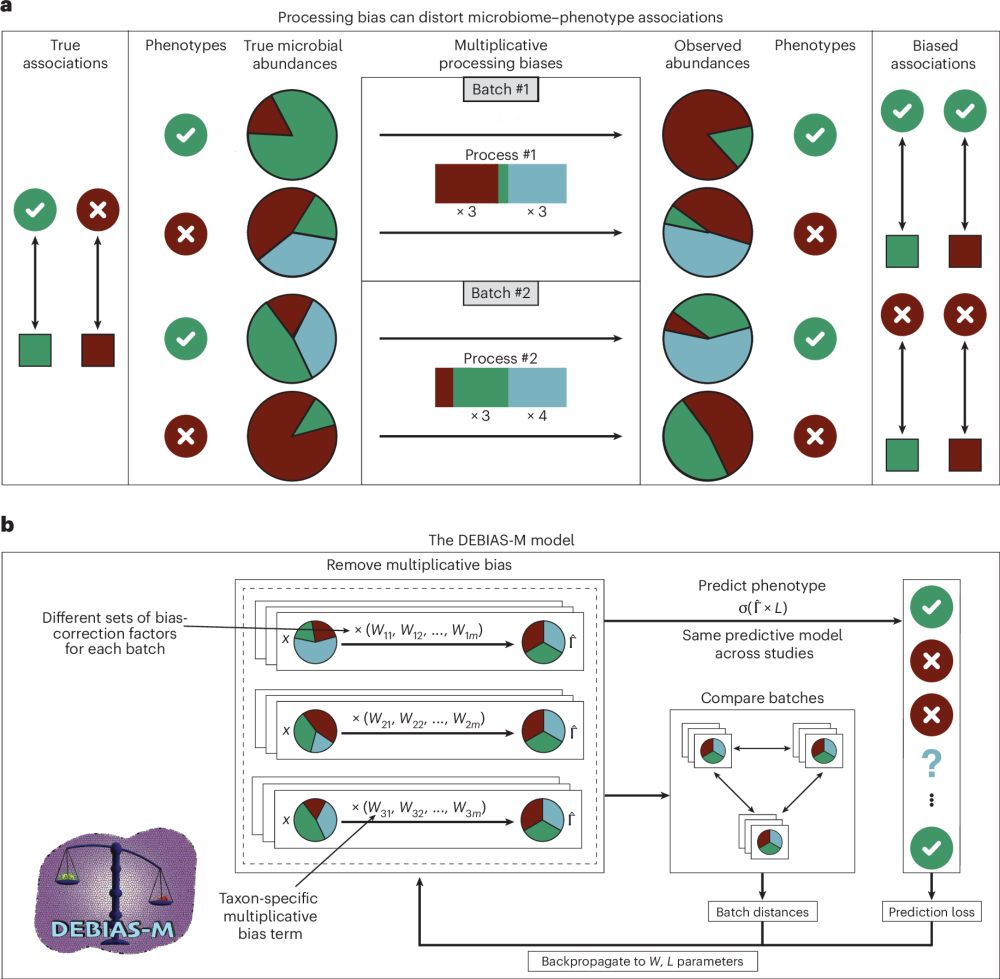
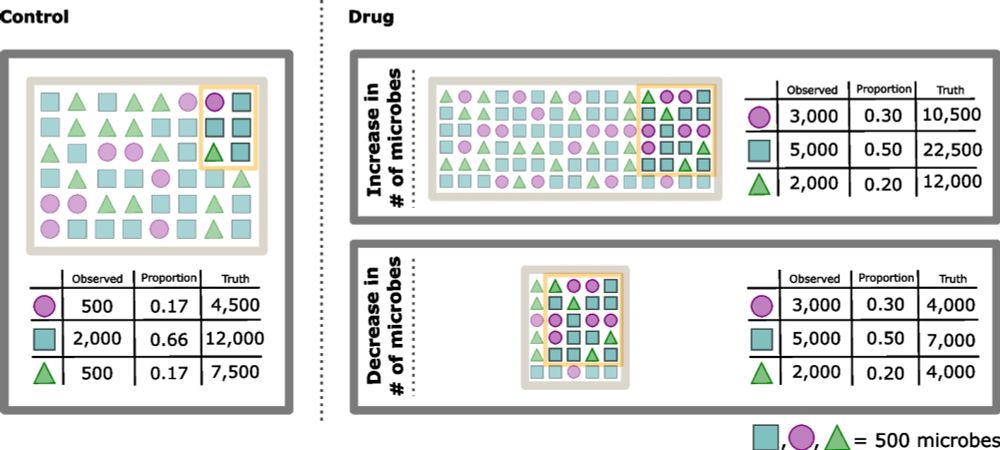
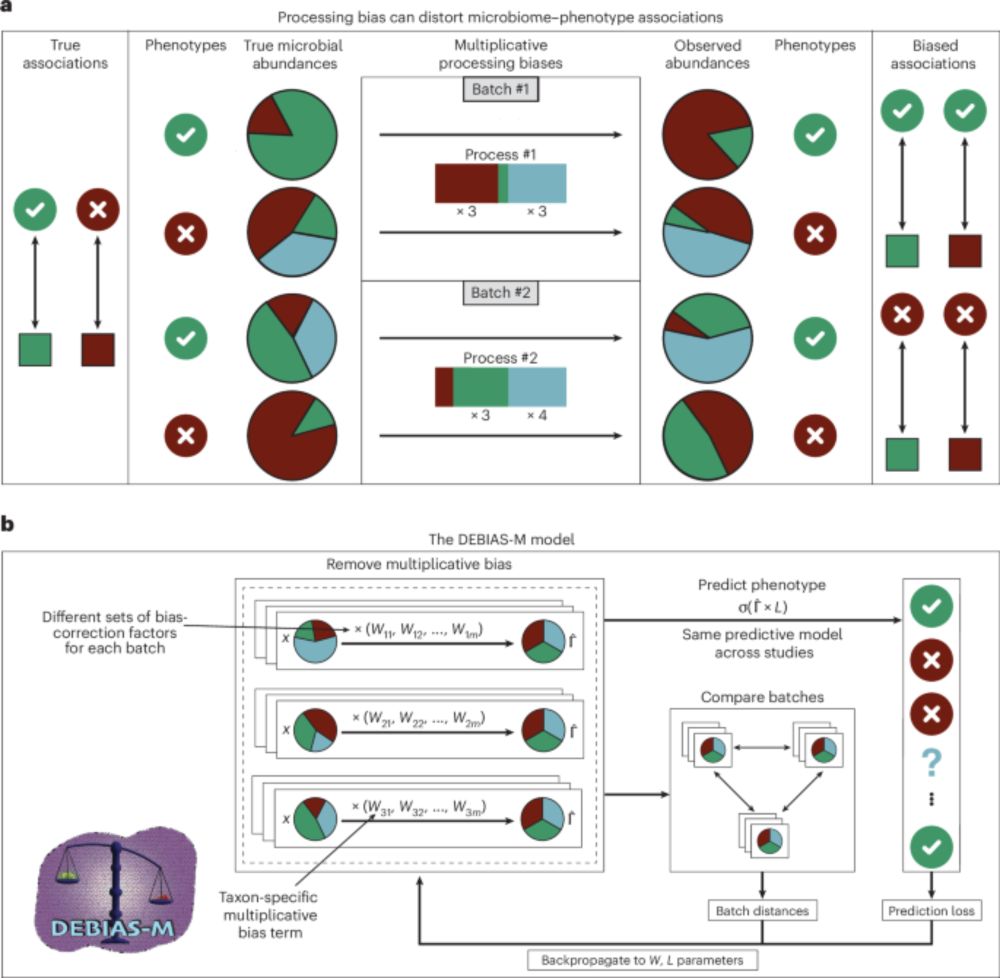
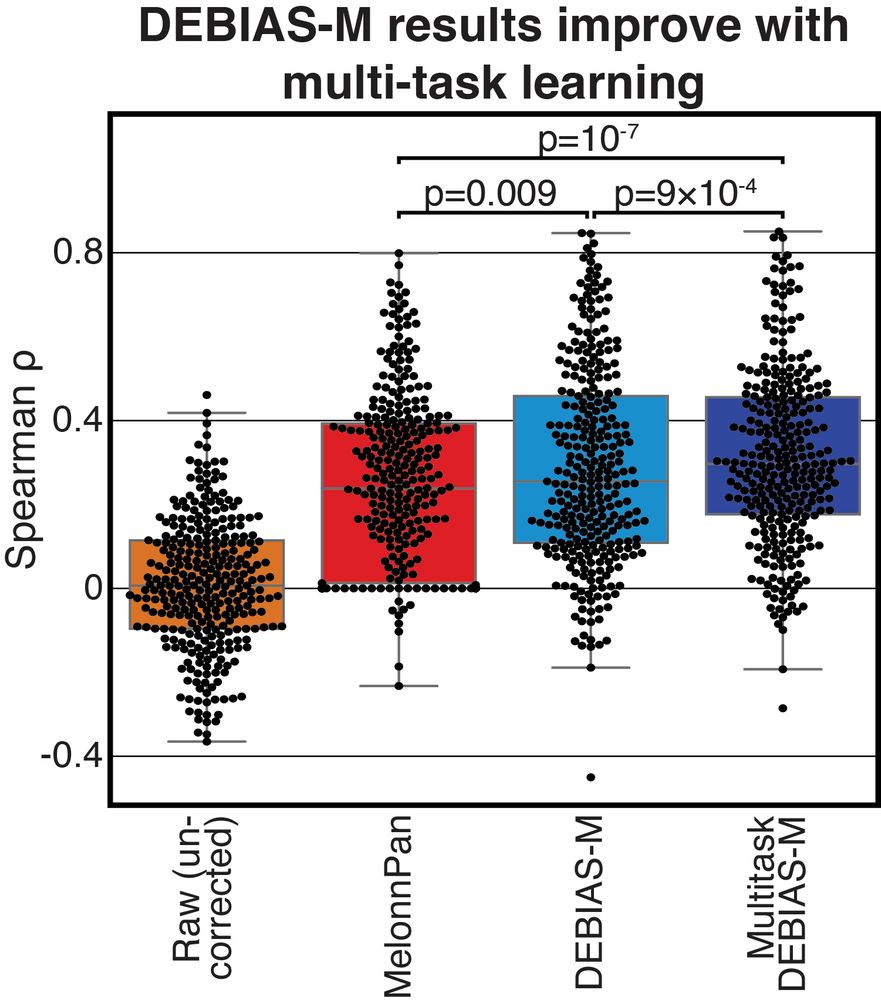
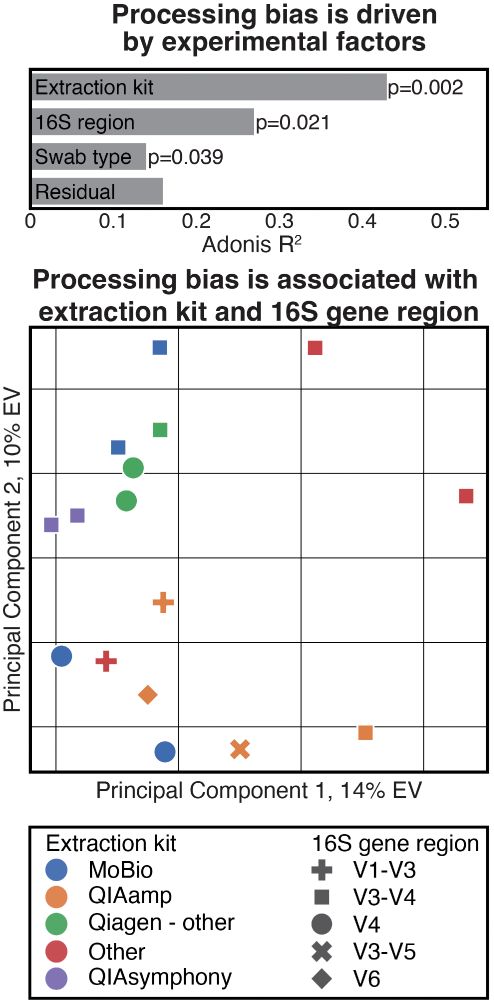
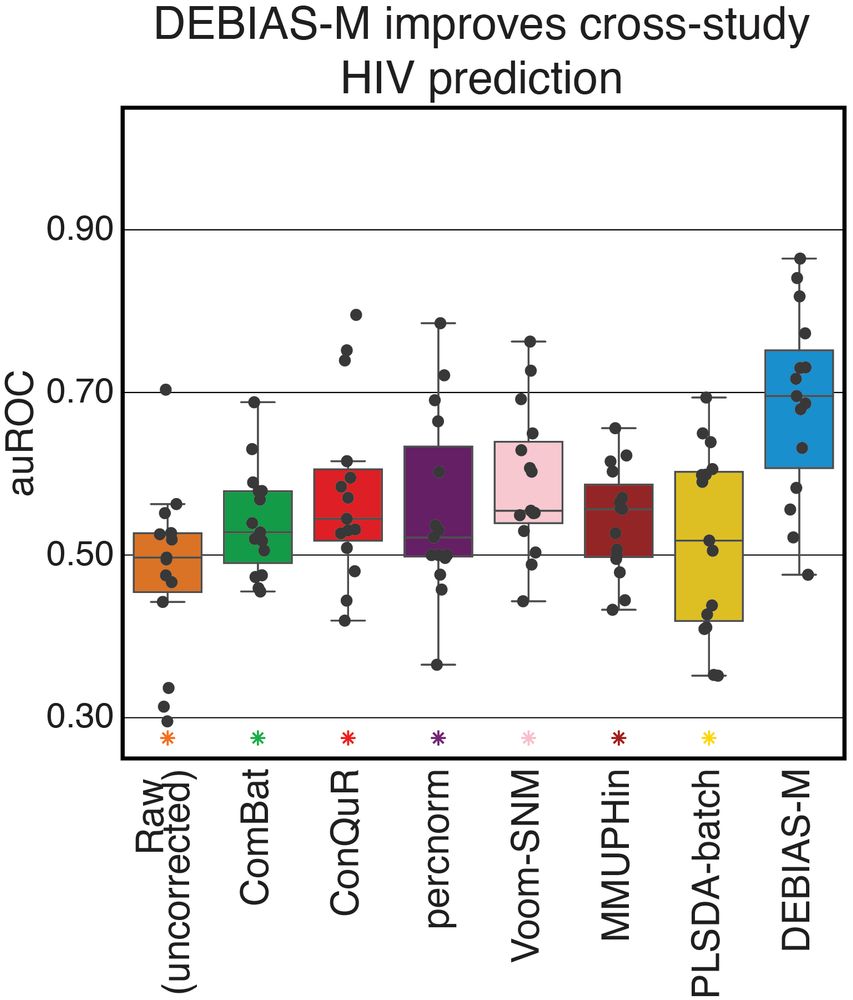
Processing-bias correction with DEBIAS-M improves cross-study generalization of microbiome-based prediction models
Nature Microbiology - DEBIAS-M corrects technical variability in microbiome data in a manner both interpretable and suitable for machine learning. In extensive benchmarks, DEBIAS-M facilitates...
www.nature.com
Tal Korem
@tkorem.bsky.social
· Mar 27
Tal Korem
@tkorem.bsky.social
· Mar 27