Bin Wan
@wanb.bsky.social
150 followers
290 following
23 posts
Computational Neurogenetics
PostDoc @ Geneva Psychiatry
PhD @mpicbs.bsky.social
Personal web: https://wanb-psych.netlify.app/
Posts
Media
Videos
Starter Packs
Pinned
Bin Wan
@wanb.bsky.social
· Jul 8
Reposted by Bin Wan
Bin Wan
@wanb.bsky.social
· Jun 23
Reposted by Bin Wan
Bin Wan
@wanb.bsky.social
· Jun 10
Reposted by Bin Wan
Nature Methods
@natmethods.nature.com
· Jun 6
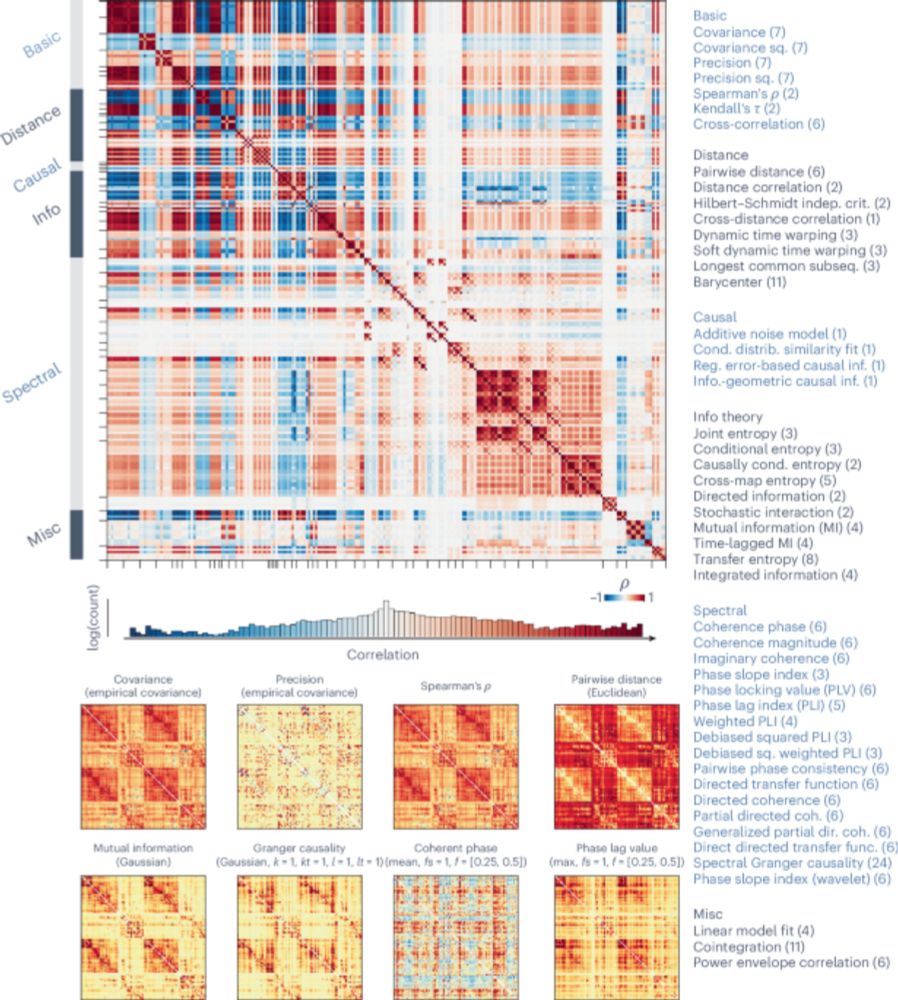
Benchmarking methods for mapping functional connectivity in the brain - Nature Methods
In this Analysis, Liu et al. benchmark more than 200 pairwise statistics for functional brain connectivity in tasks such as hub mapping, distance relationships, structure–function coupling and behavio...
www.nature.com
Reposted by Bin Wan
Reposted by Bin Wan
Reposted by Bin Wan
Bin Wan
@wanb.bsky.social
· May 19
Reposted by Bin Wan
Reposted by Bin Wan
Reposted by Bin Wan
Bin Wan
@wanb.bsky.social
· Apr 5
Bin Wan
@wanb.bsky.social
· Mar 5

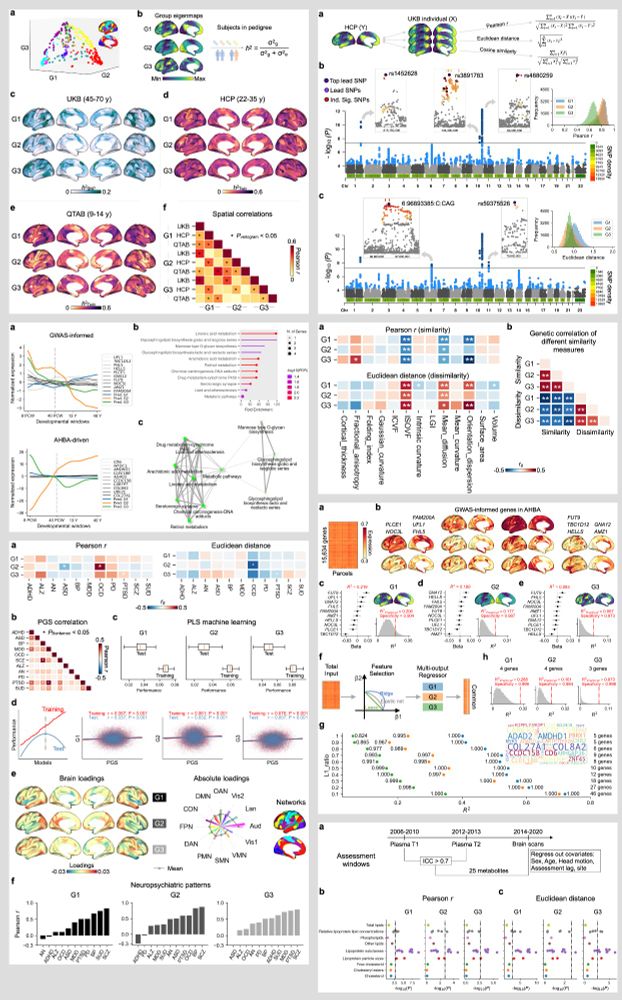
Genetic, transcriptomic, metabolic, and neuropsychiatric underpinnings of cortical functional gradients
Functional gradients capture the organization of functional activity in the cerebral cortex, delineating transitions from sensory to higher-order association areas. While group-level gradient patterns...
www.medrxiv.org
Bin Wan
@wanb.bsky.social
· Mar 5

Genetic, transcriptomic, metabolic, and neuropsychiatric underpinnings of cortical functional gradients
Functional gradients capture the organization of functional activity in the cerebral cortex, delineating transitions from sensory to higher-order association areas. While group-level gradient patterns...
www.medrxiv.org
Bin Wan
@wanb.bsky.social
· Mar 5
Bin Wan
@wanb.bsky.social
· Mar 5
Reposted by Bin Wan
James Pang
@jchrispang.bsky.social
· Feb 3

Geometric influences on the regional organization of the mammalian brain
The mammalian brain is comprised of anatomically and functionally distinct regions. Substantial work over the past century has pursued the generation of ever-more accurate maps of regional boundaries,...
www.biorxiv.org