Wei-Ting Lu
@weitinglu.bsky.social
180 followers
490 following
23 posts
Tea addict and scientist, in that order. Incoming Group Leader, Dept of Oncology, University of Oxford. All views my own.
Posts
Media
Videos
Starter Packs
Reposted by Wei-Ting Lu
Reposted by Wei-Ting Lu
Cancer Research UK
@cancerresearchuk.org
· Apr 27
Reposted by Wei-Ting Lu
Esashi Lab
@esashilab.bsky.social
· Feb 25
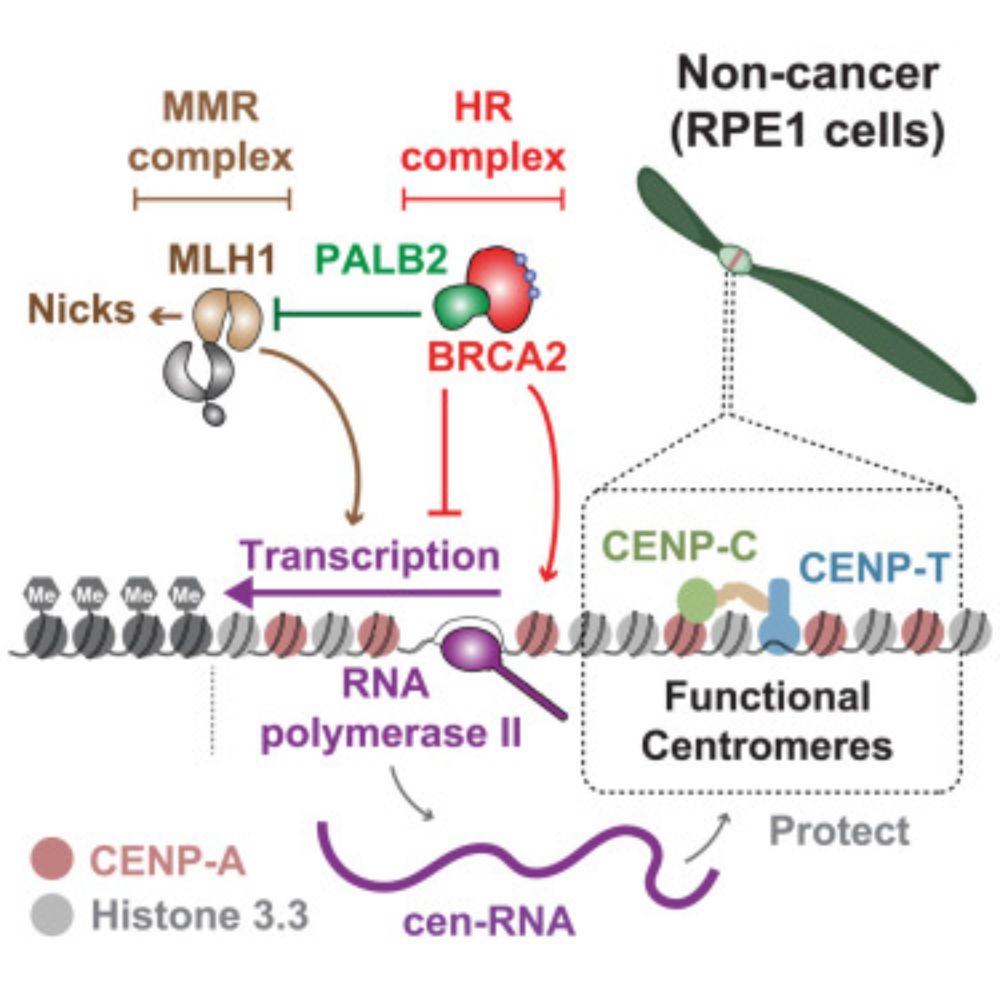
The homologous recombination factors BRCA2 and PALB2 interplay with mismatch repair pathways to maintain centromere stability and cell viability
Graham et al. demonstrate that BRCA2 and PALB2 protect centromere integrity in non-cancerous
cells through distinct mechanisms, partly counteracting the mismatch repair factor
MLH1. In cancer cells, B...
www.cell.com
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 30
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 17
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 17
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 17
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 3
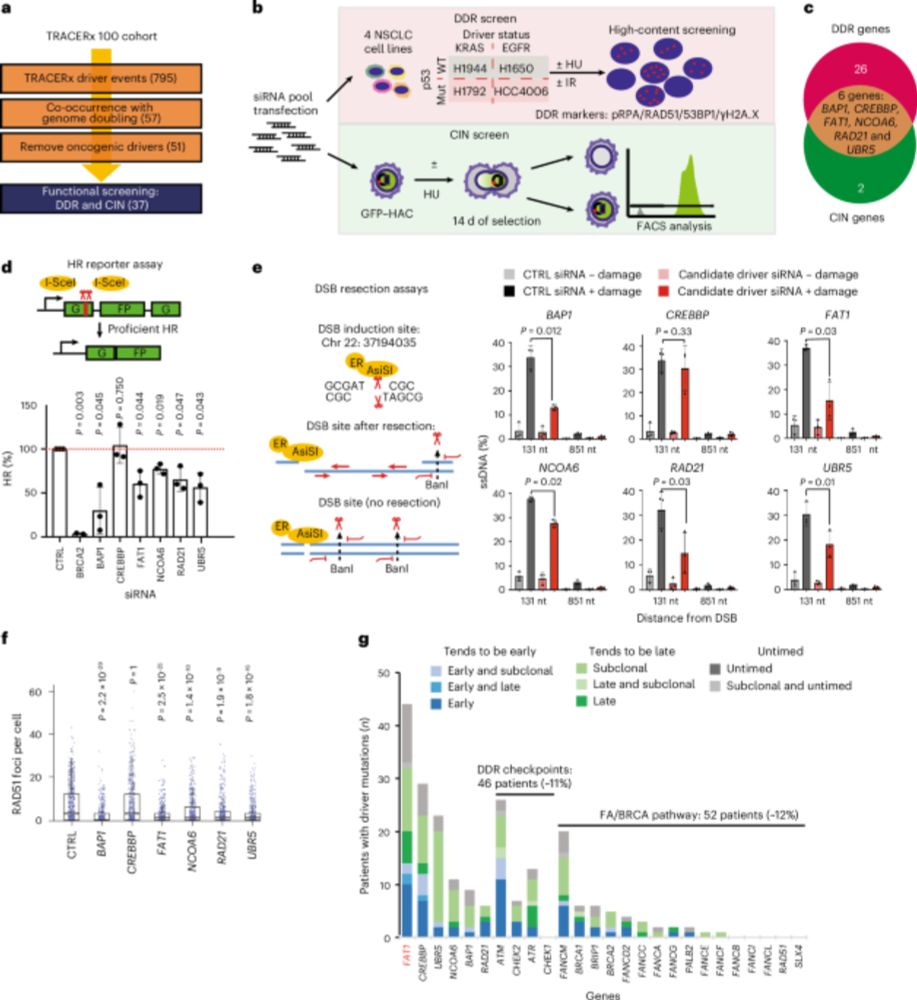
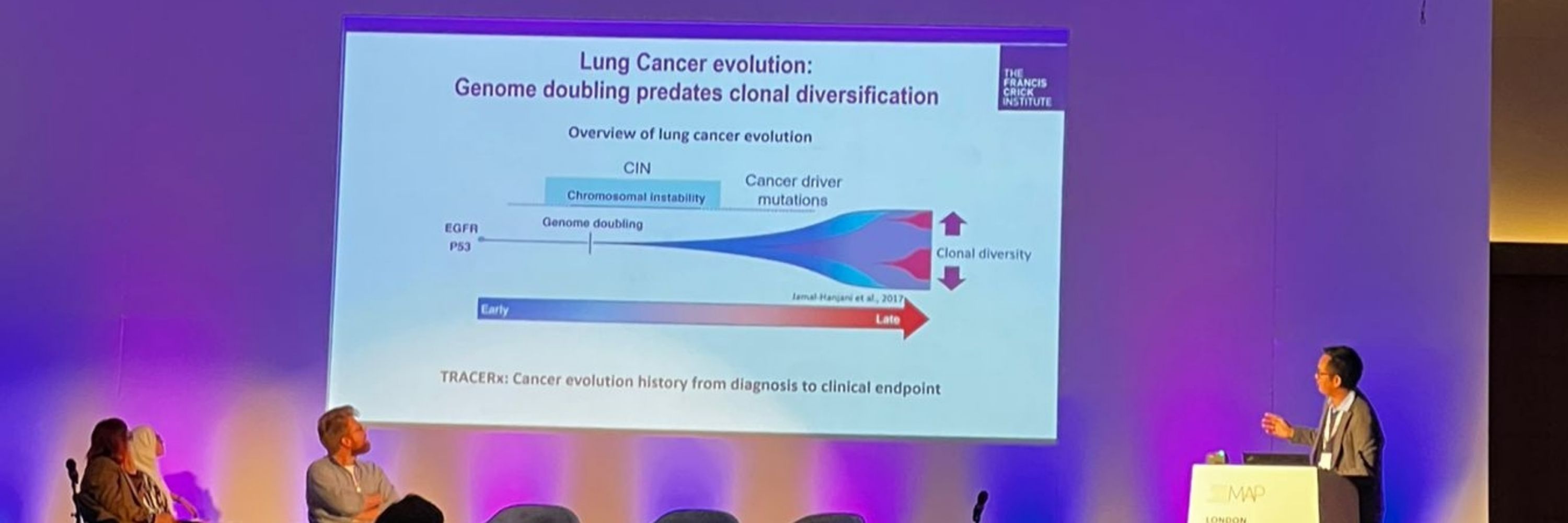
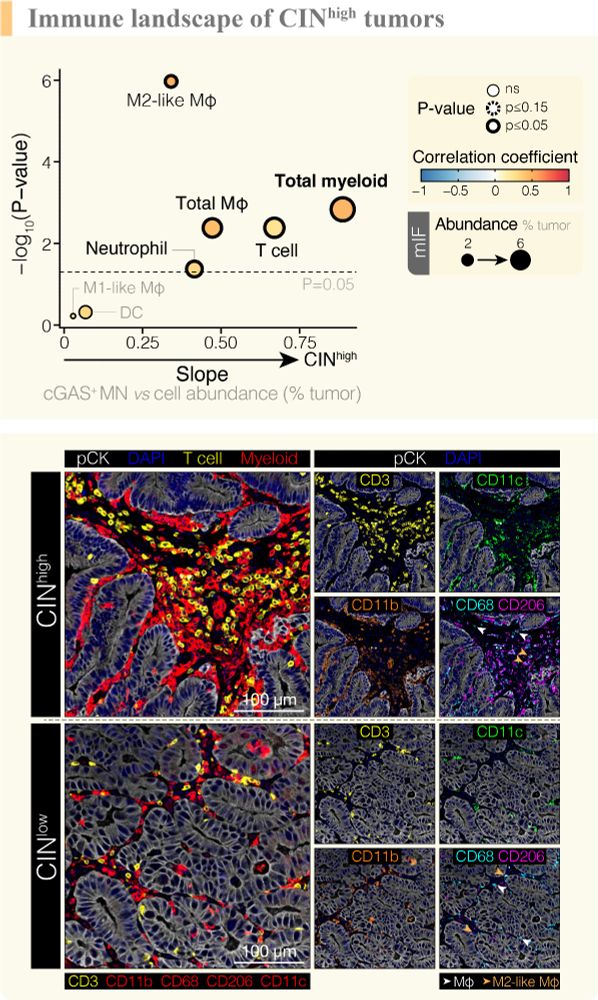
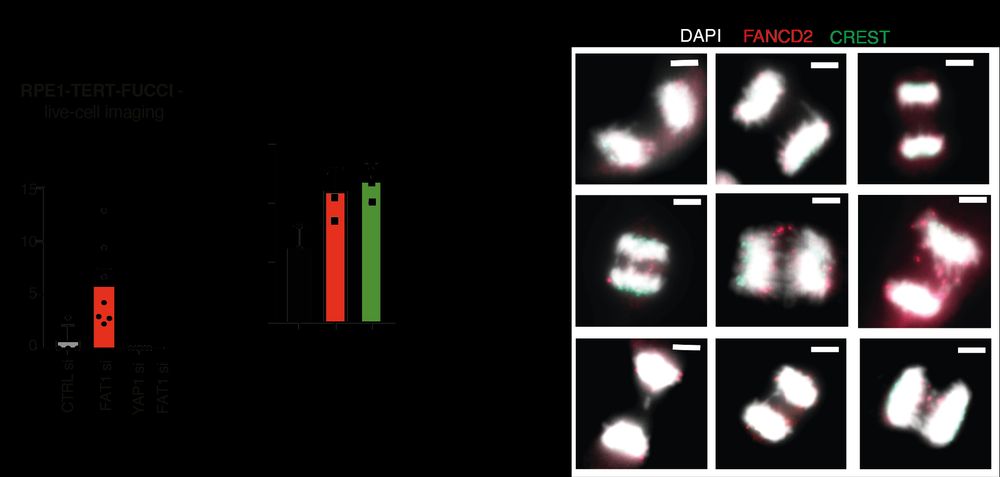
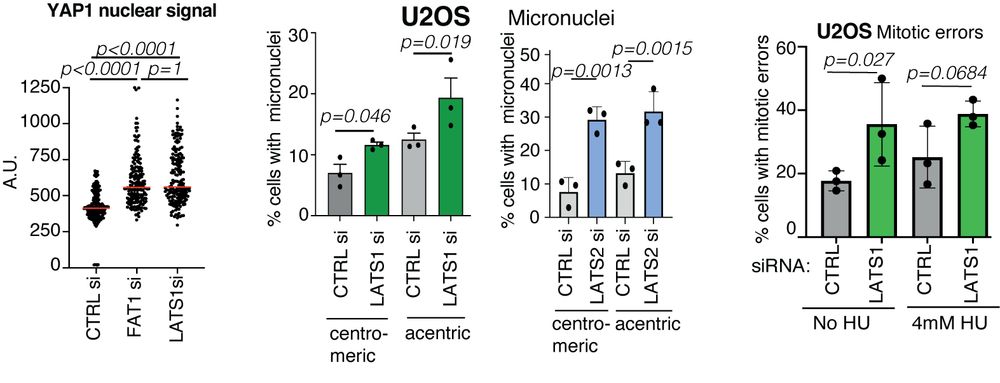
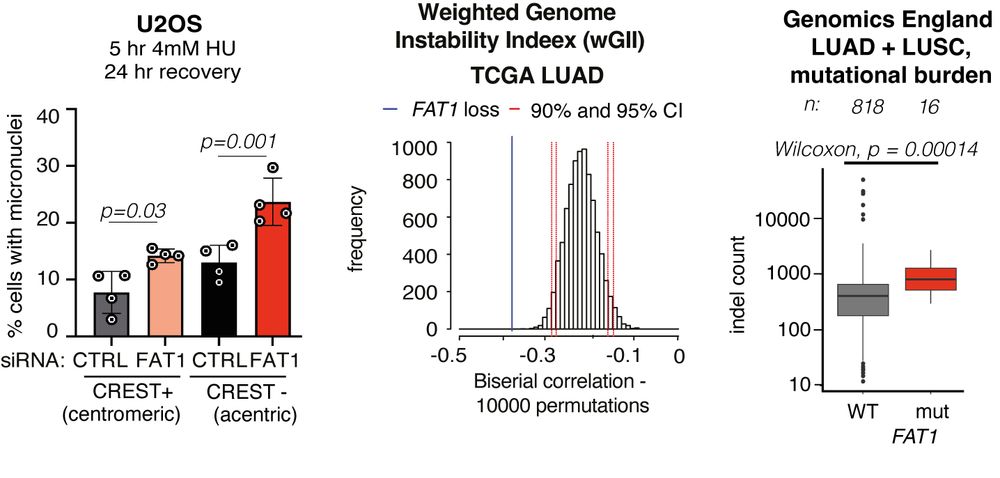
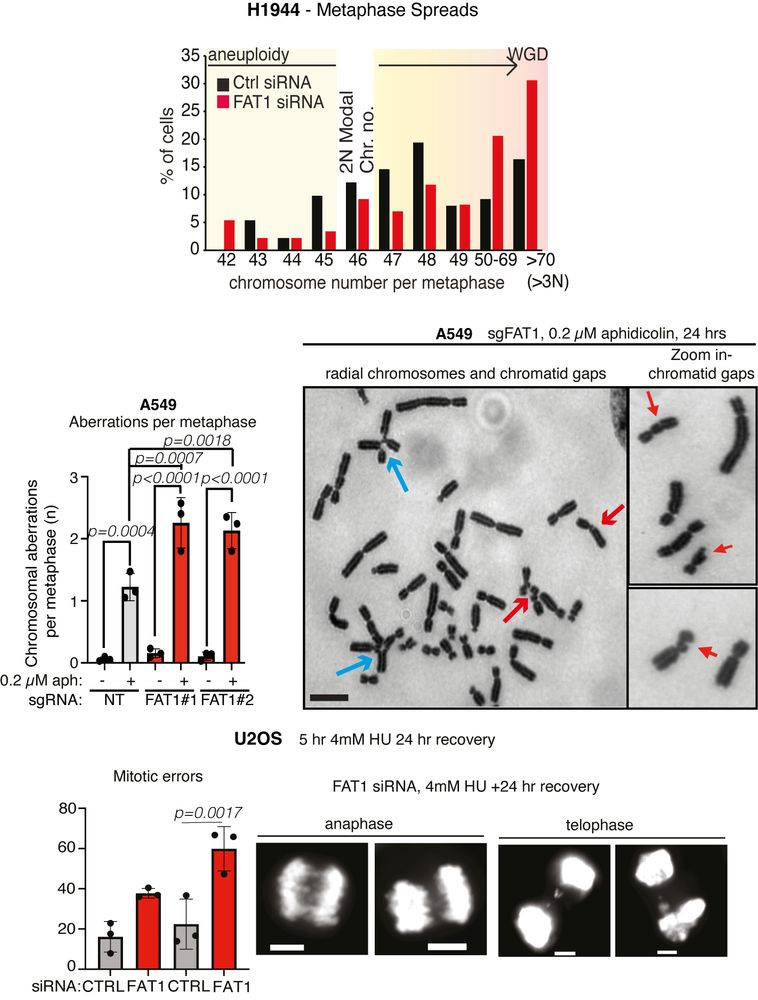
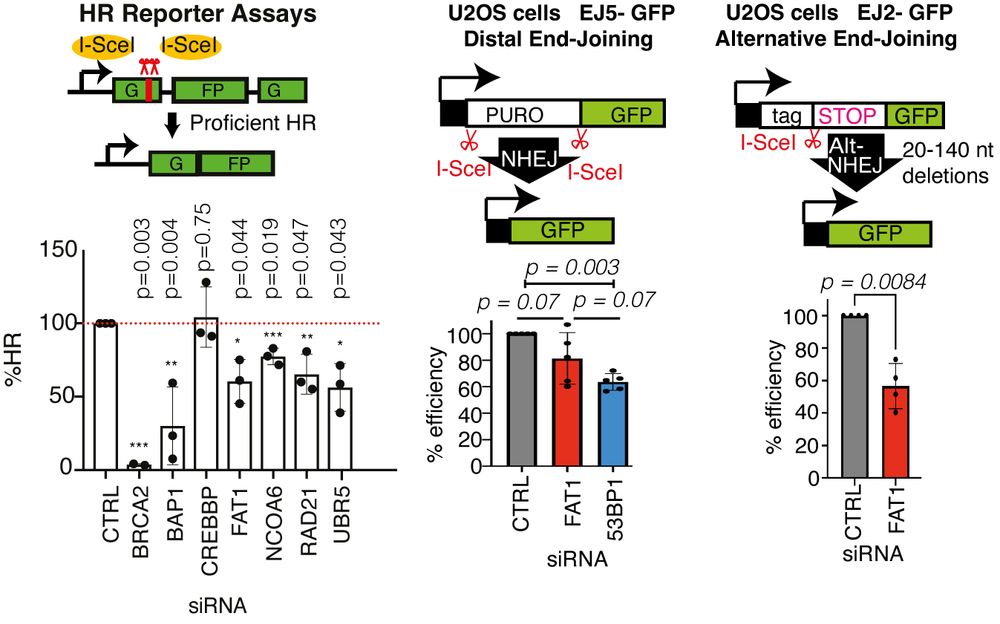
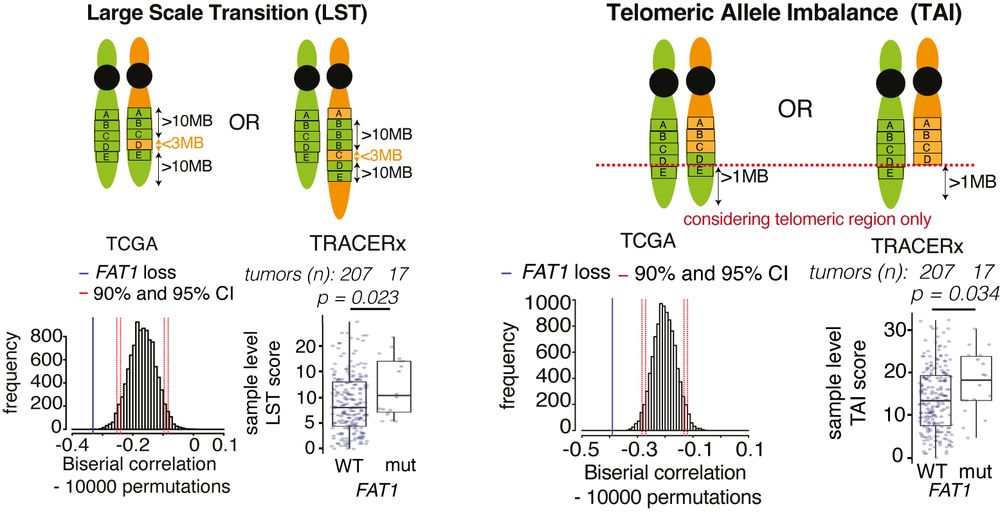
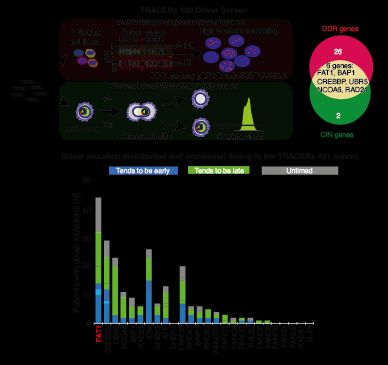
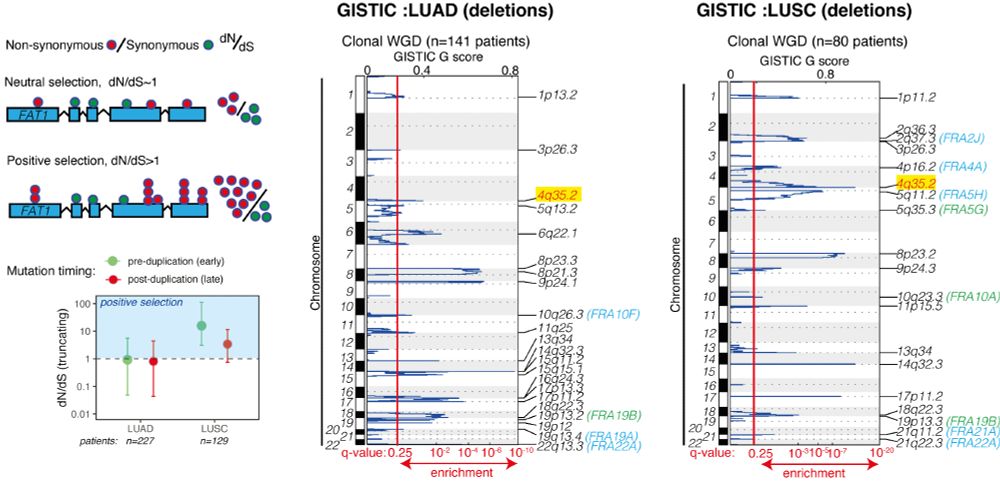
TRACERx analysis identifies a role for FAT1 in regulating chromosomal instability and whole-genome doubling via Hippo signalling - Nature Cell Biology
Lu et al. perform systematic functional analyses using data from the TRACERx cohort of patients with non-small-cell lung cancer and delineate how FAT1 regulates homologous recombination repair, chromo...
www.nature.com
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 3
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 3
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 3
Wei-Ting Lu
@weitinglu.bsky.social
· Jan 3