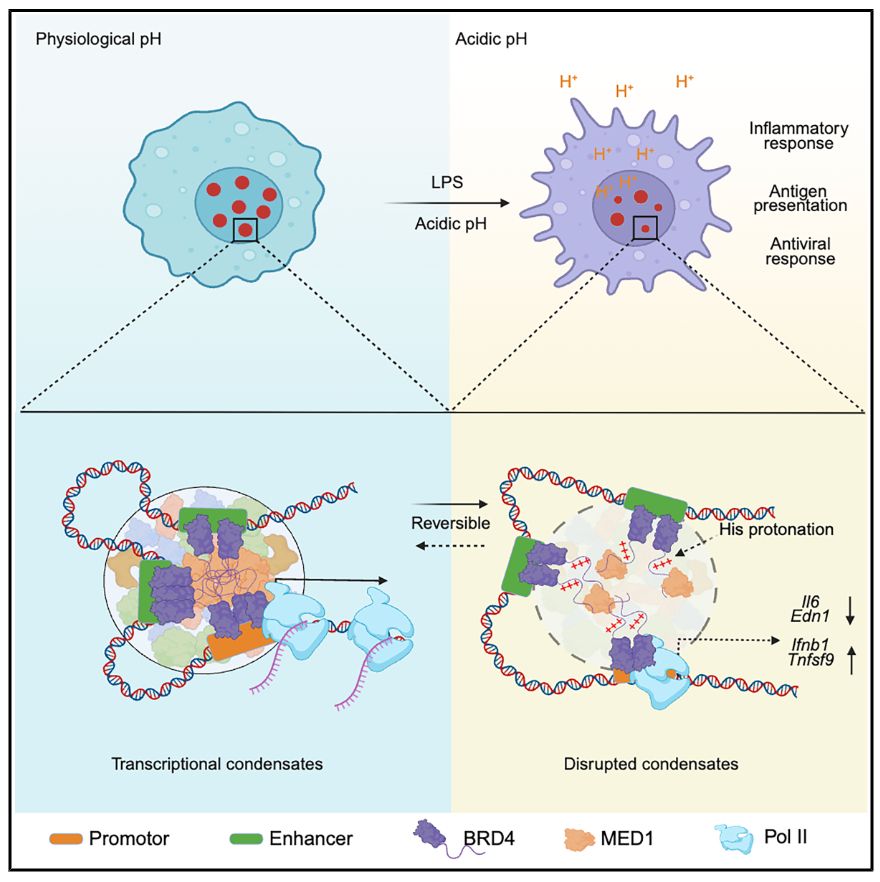
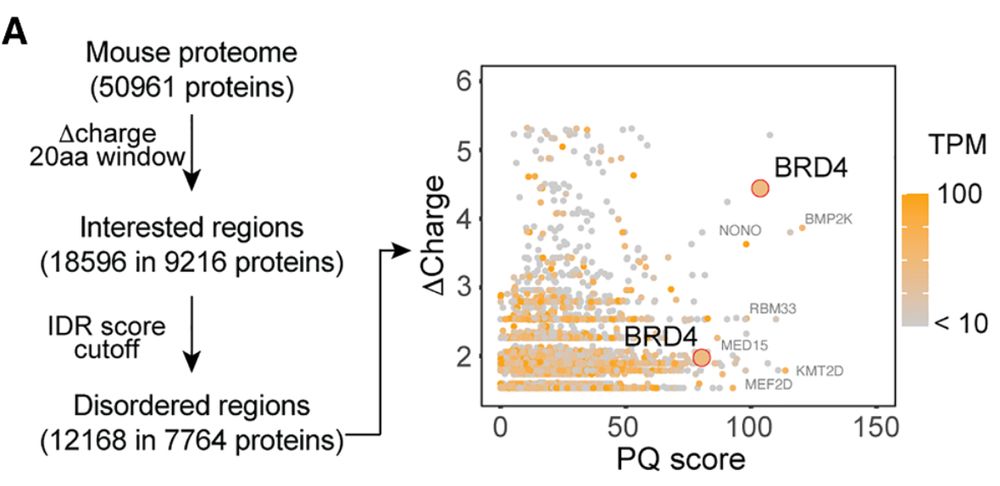
Xu Zhou lab @ BCH & HMS
@xuzhoulab.bsky.social
440 followers
640 following
47 posts
AP @BostonChildrens @harvardmed AM @broadinstitute
inflammation, tissue biology, quant.&systems immunology #1stGen #ImmigrantScientists #NewPI
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Xu Zhou lab @ BCH & HMS