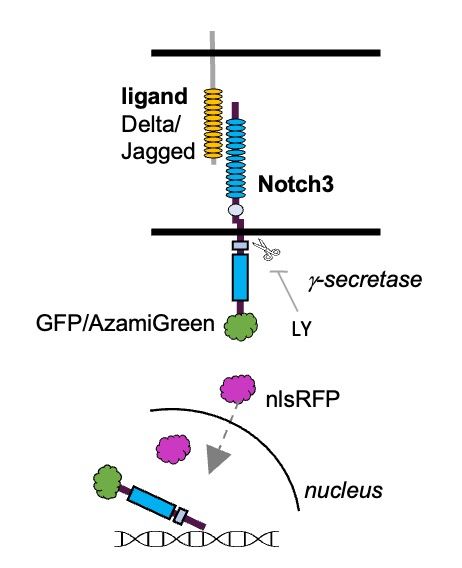
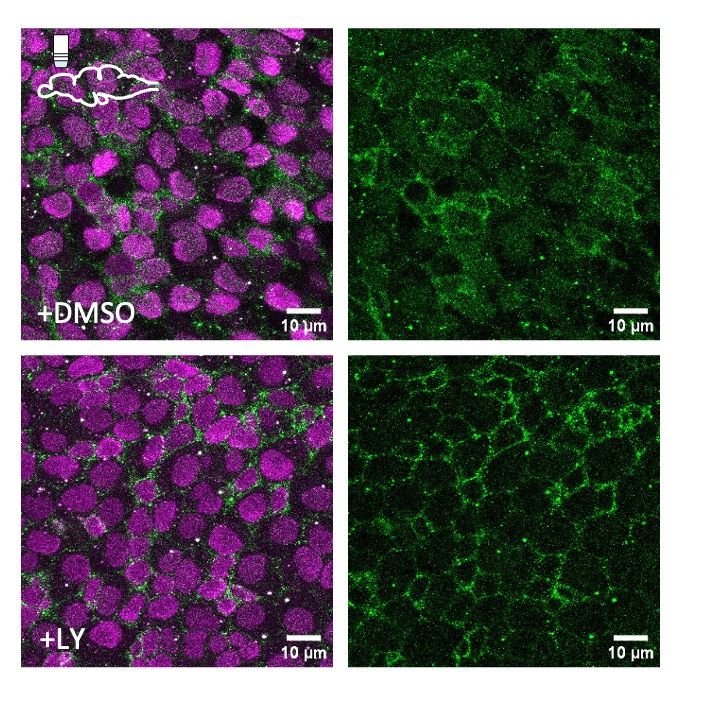
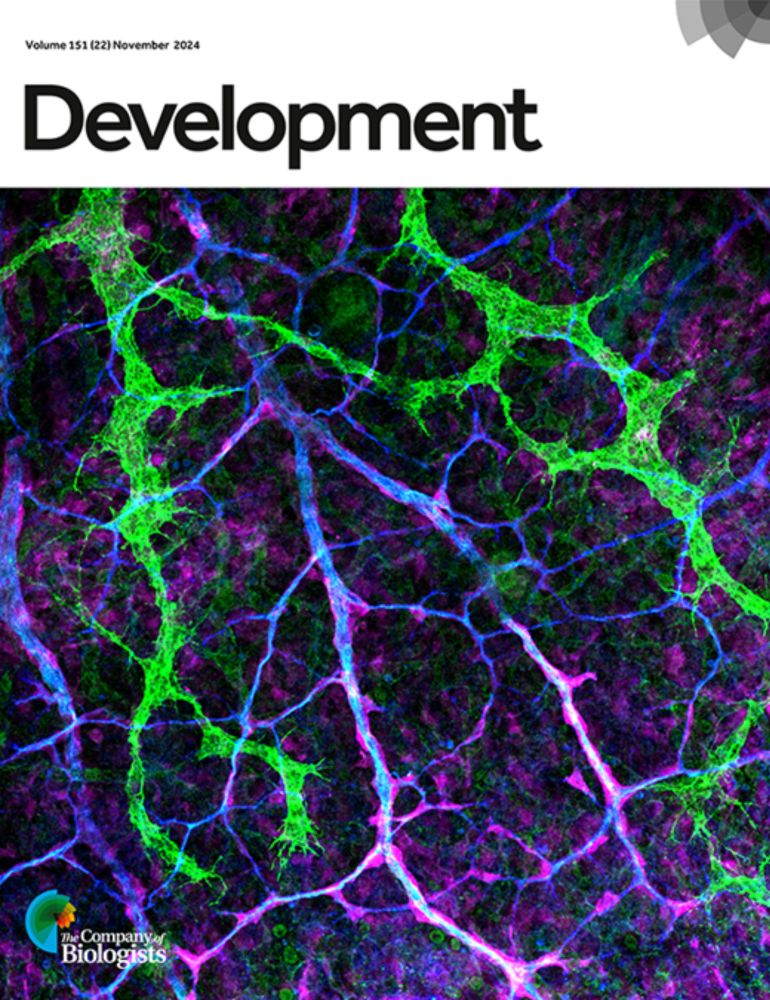
Posts
Media
Videos
Starter Packs
Reposted by Zenlabpasteur.bsky.social
Reposted by Zenlabpasteur.bsky.social
Reposted by Zenlabpasteur.bsky.social
Reposted by Zenlabpasteur.bsky.social