What happens after variant calling is often ad hoc.
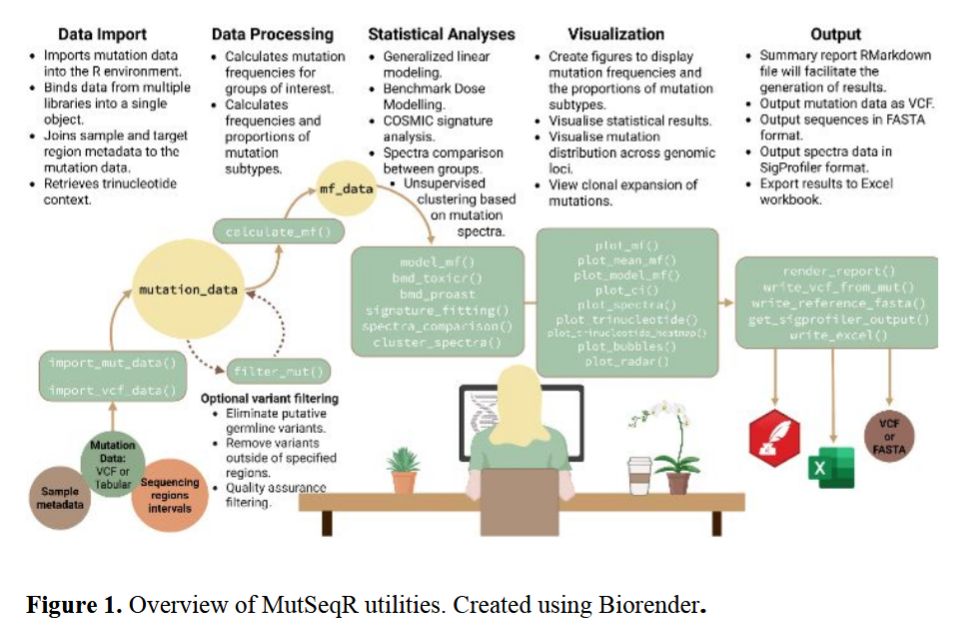
MutSeqR picks up there for mutation frequency modeling, spectrum analysis, & benchmark dose estimation across ECS platforms.
More detail here:
🔗 blog.fulcrumgenomics.com/p/mutseqr-op...

What happens after variant calling is often ad hoc.
MutSeqR picks up there for mutation frequency modeling, spectrum analysis, & benchmark dose estimation across ECS platforms.
More detail here:
🔗 blog.fulcrumgenomics.com/p/mutseqr-op...
📄 Paper: doi.org/10.1093/bioa...
💻 Code: github.com/EHSRB-BSRSE-Bioinformatics/MutSeqR
#OpenScience #Genomics
📄 Paper: doi.org/10.1093/bioa...
💻 Code: github.com/EHSRB-BSRSE-Bioinformatics/MutSeqR
#OpenScience #Genomics
It brings much-needed standardization to genetic toxicology research.
🧬 🖥️ blog.fulcrumgenomics.com/p/mutseqr-op...

It brings much-needed standardization to genetic toxicology research.
🧬 🖥️ blog.fulcrumgenomics.com/p/mutseqr-op...
→ variant filtering
→ mutation frequency + dose-response modeling
→ benchmark dose estimation
→ spectrum + signature analysis
→ reproducible visualization tools
Openly available at 🔗 github.com/EHSRB-BSRSE-Bioinformatics/MutSeqR

→ variant filtering
→ mutation frequency + dose-response modeling
→ benchmark dose estimation
→ spectrum + signature analysis
→ reproducible visualization tools
Openly available at 🔗 github.com/EHSRB-BSRSE-Bioinformatics/MutSeqR
Developed with Health Canada & U Ottawa, it helps bring consistency to genetic safety research.
🔗 doi.org/10.1093/bioa...
#Genomics #OpenSource #MutSeqR 🧬🖥️

Developed with Health Canada & U Ottawa, it helps bring consistency to genetic safety research.
🔗 doi.org/10.1093/bioa...
#Genomics #OpenSource #MutSeqR 🧬🖥️

Full article available: https://doi.org/10.1093/bioadv/vbaf265
Full article available: https://doi.org/10.1093/bioadv/vbaf265

