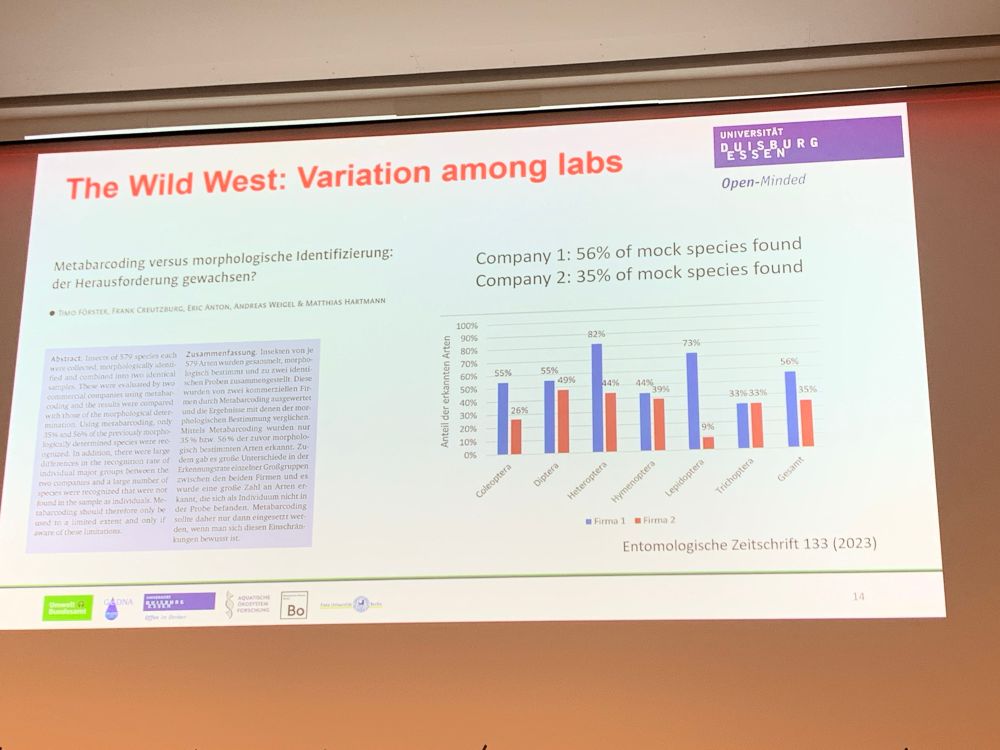
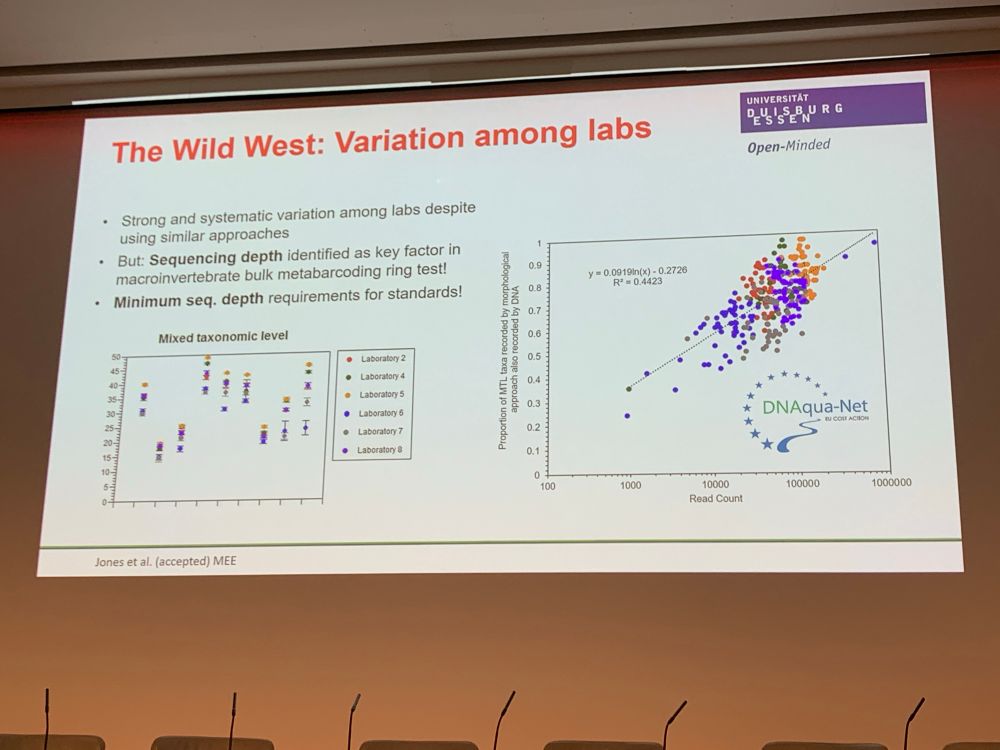
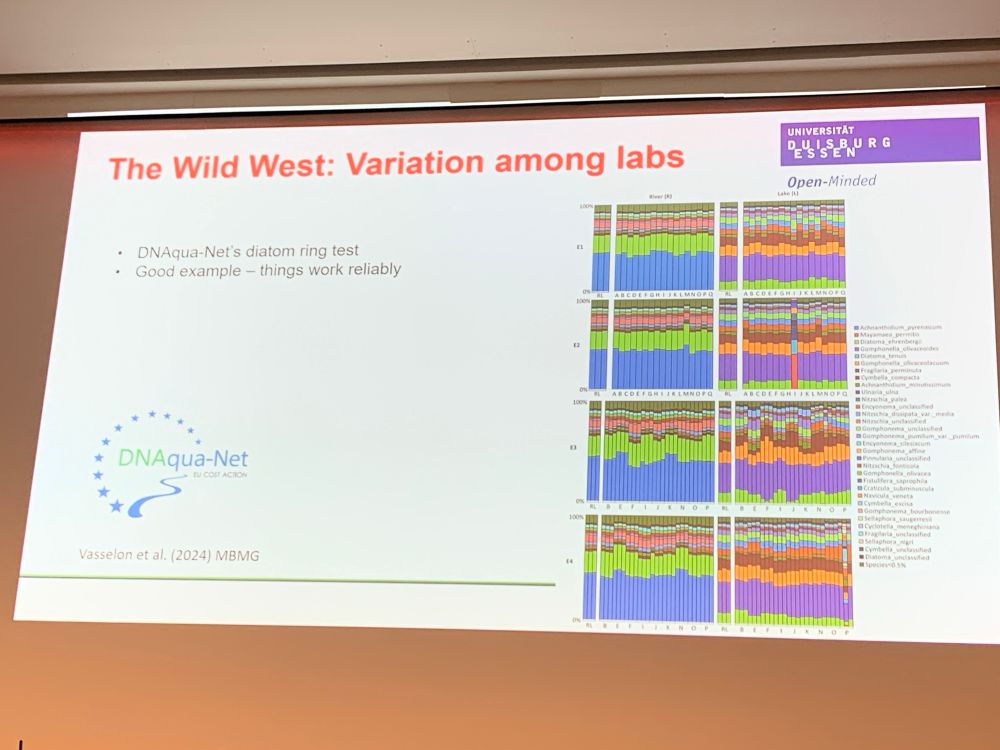
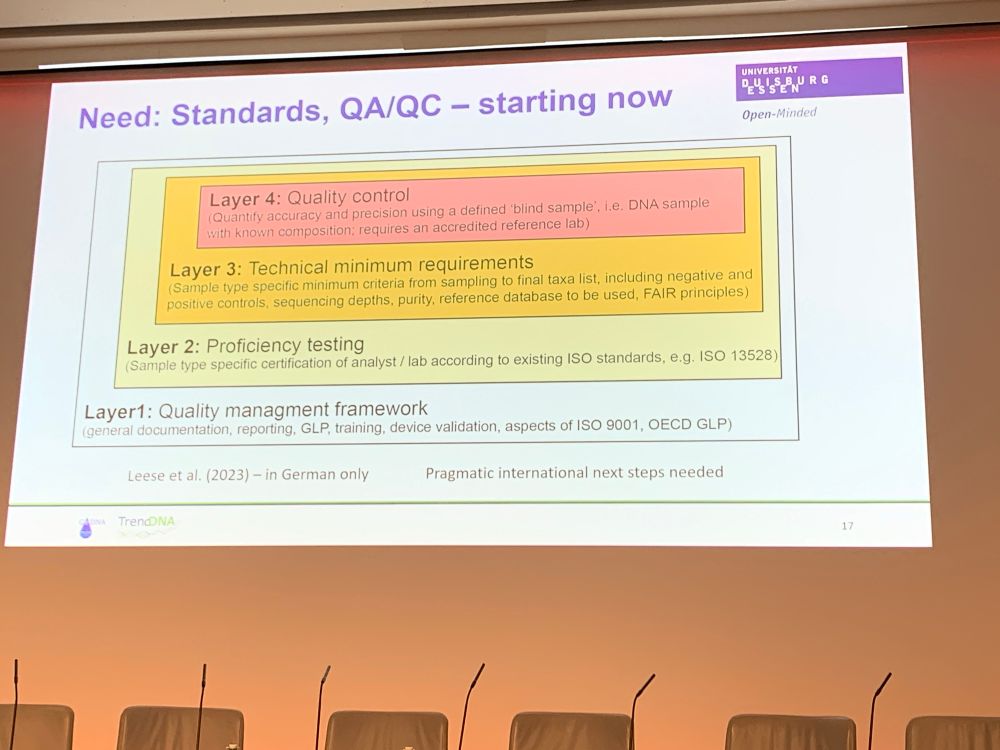
by Florian Altermatt — Reposted by: Florian Leese, Julie L. Lockwood, Florian Altermatt
Now results out in 2 #parallel #preprints @biorxiv-ecology.bsky.social
#aquatic #biodiversity #meta-analysis
1/6🧵

Long #Day1 today as we sampled until 19:30. Motivation: 10/10. Tomorrow: to the lab! @unidue.bsky.social @sgn.one




Reposted by: Florian Leese, Daniel Hering

by Doerthe Tetzlaff — Reposted by: Florian Leese
by Florian Leese — Reposted by: Doerthe Tetzlaff
Time for inter- & transdisciplinary discussions. Also: Award ceremony of new junior #research group
water-research-horizon.de/wrhc.html RT


Reposted by: Florian Leese, Doerthe Tetzlaff
🔗 doi.org/10.3897/mbmg...

Reposted by: Florian Leese
Freely available at: dx.doi.org/10.1002/ece3...

Reposted by: Florian Leese
👇 sgn.one/8x2

Reposted by: Florian Leese
📅Save the dates: 14-18 July
🔗For more information: www.physalia-courses.org/courses-work...

Reposted by: Florian Leese
Targeting the right sequences can overcome nontarget amplification
👉 bit.ly/4dC2rlT
#science #water

Reposted by: Florian Leese
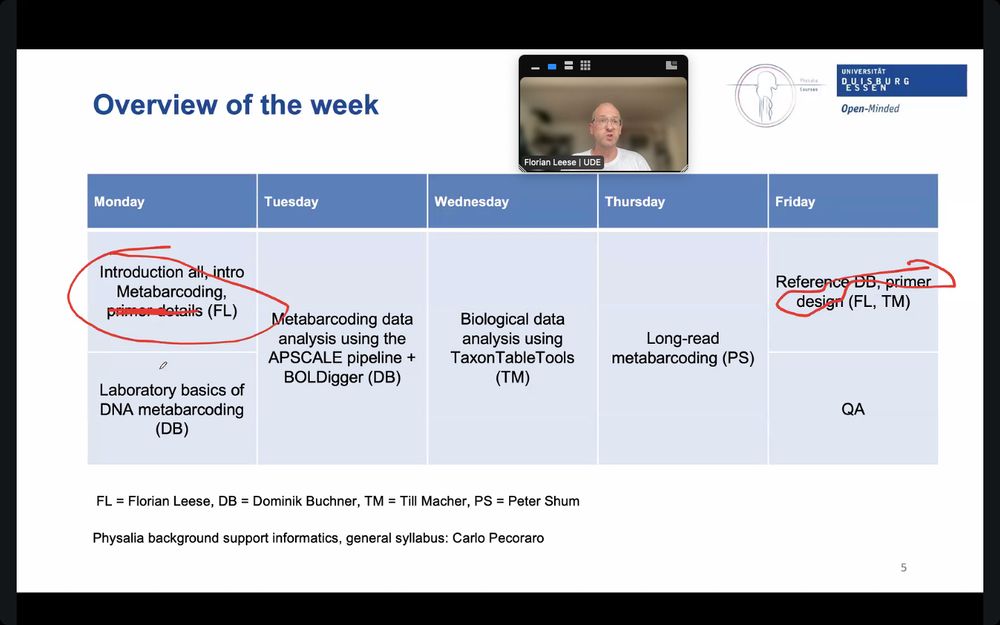
We explored amazing user-friendly tools like BolDigger, Aspcale, and TaxonTable, had a great intro to @nanoporetech.com data, and wrapped up with an inspiring open discussion full of ideas and projects.

#freshwater #biodiversity #multiplestressors
Reposted by: Florian Leese, Daniel Hering
Ich freue mich sehr, dass unser Buch „Von Angesicht zu Angesicht“ dabei ist!
Ihr könnt uns sowie unsere Arbeit, mit der wir für biologische Vielfalt begeistern, mit eurer Stimme in der Kategorie „Ästhetik“ unterstützen: www.konradin-service.de/umfrage/inde...

Reposted by: Florian Leese
View post
Reposted by: Florian Leese
Finally, thank you @leeselab.bsky.social for sitting in the thesis committee! 🫶

Reposted by: Florian Leese

by Florian Leese — Reposted by: Daniel Hering
Really happy & grateful to work here with such a great bunch of talented & nice people - connected to our collaborators around the 🌎. Life could be worse. Weiter geht‘s. ☀️Maybe more lab work next 🔟 ?




While I personally don’t care much about these metrics I know they are really relevant for many institutions. Thus: very happy about this!

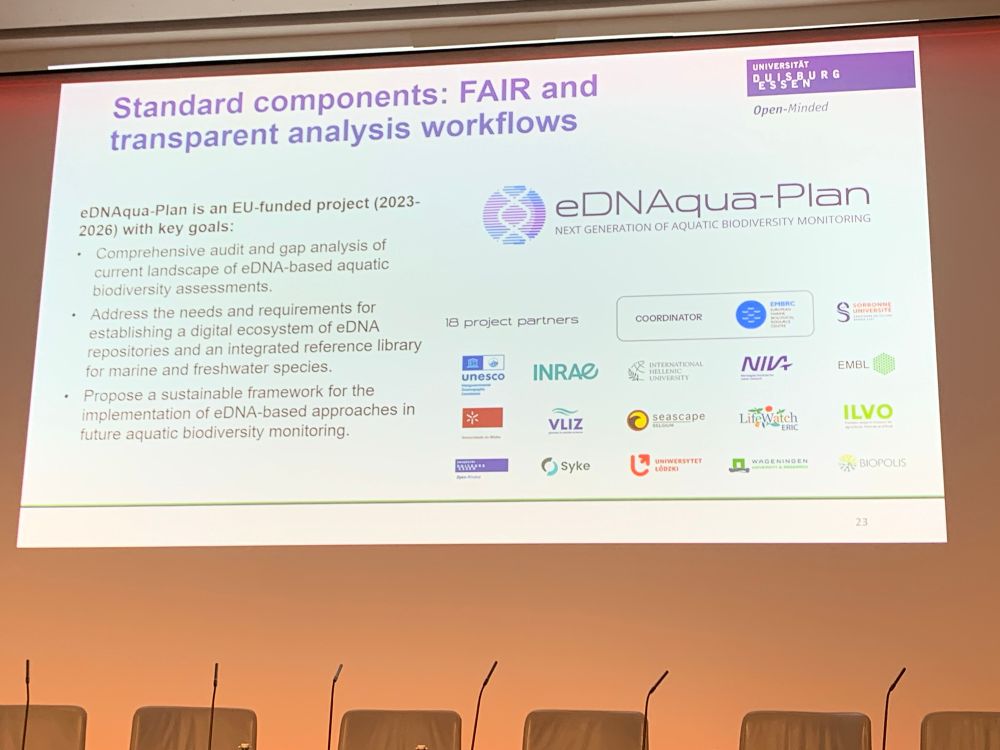
Kommt vorbei, meldet euch an: www.dgl-jahrestagungen.de/phone/index....
#Vortrag #Poster #Tagung #Limnologie #omics #eDNA #Monitoring


Reposted by: Florian Leese
shorturl.at/wRW4T Till & @shump.bsky.social
Get hands-on experience analysing Illumina & @nanoporetech.com data & learn how to run metabarcoding pipelines from raw sequences to ecological insights

Reposted by: Florian Leese
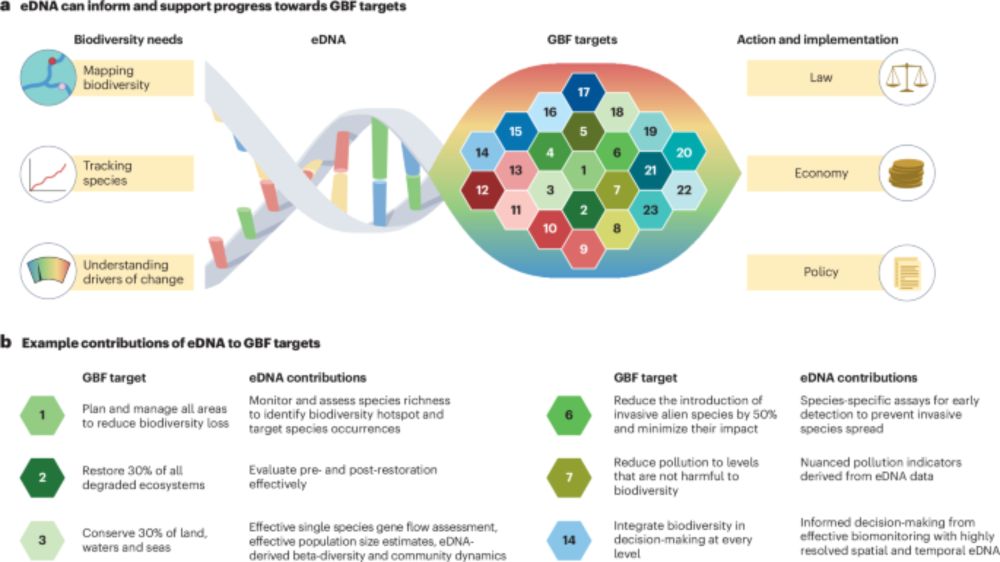
Web link: go.nature.com/4mv2h4W
Readcube: rdcu.be/eh0uC
Reposted by: Florian Leese
Reposted by: Florian Leese, Daniel Hering



Reposted by: Florian Leese



Reposted by: Florian Leese