
Chris Anderton ⚜️
@anderton.bsky.social
170 followers
180 following
10 posts
Spatial metabolomics of everything 👨🦲🌲🌱 🦠 🍄, advancing mass spec imaging technology and molecular cartography(➕I ❤️🏎️🏀🏈)
Posts
Media
Videos
Starter Packs
Reposted by Chris Anderton ⚜️
Reposted by Chris Anderton ⚜️
Reposted by Chris Anderton ⚜️
Chris Anderton ⚜️
@anderton.bsky.social
· Mar 27

Imaging and spatially resolved mass spectrometry applications in nephrology - Nature Reviews Nephrology
In this Review, the authors discuss mass spectrometry (MS) imaging and spatially resolved MS approaches that are being employed in nephrology applications. They also highlight emerging MS methods and ...
www.nature.com
Chris Anderton ⚜️
@anderton.bsky.social
· Feb 28

The Impact of the Mass Analyzer and Tissue Section Thickness on Spatial N-Glycomics with MALDI-MSI
We compared matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI) spatial N-glycomics data sets from Fourier-transform ion cyclotron resonance (FTICR) and orthogonal accele...
pubs.acs.org
Reposted by Chris Anderton ⚜️
Reposted by Chris Anderton ⚜️
Chris Anderton ⚜️
@anderton.bsky.social
· Dec 22
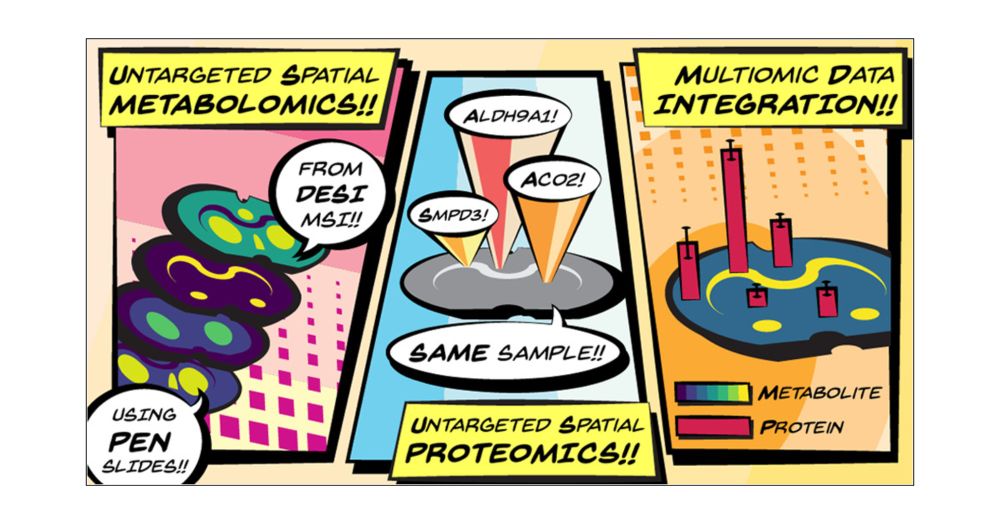
Untargeted Spatial Metabolomics and Spatial Proteomics on the Same Tissue Section
An increasing number of spatial multiomic workflows have recently been developed. Some of these approaches have leveraged initial mass spectrometry imaging (MSI)-based spatial metabolomics to inform the region of interest (ROI) selection for downstream spatial proteomics. However, these workflows have been limited by varied substrate requirements between modalities or have required analyzing serial sections (i.e., one section per modality). To mitigate these issues, we present a new multiomic workflow that uses desorption electrospray ionization (DESI)-MSI to identify representative spatial metabolite patterns on-tissue prior to spatial proteomic analyses on the same tissue section. This workflow is demonstrated here with a model mammalian tissue (coronal rat brain section) mounted on a poly(ethylene naphthalate)-membrane slide. Initial DESI-MSI resulted in 160 annotations (SwissLipids) within the METASPACE platform (≤20% false discovery rate). A segmentation map from the annotated ion images informed the downstream ROI selection for spatial proteomics characterization from the same sample. The unspecific substrate requirements and minimal sample disruption inherent to DESI-MSI allowed for an optimized, downstream spatial proteomics assay, resulting in 3888 ± 240 to 4717 ± 48 proteins being confidently directed per ROI (200 μm × 200 μm). Finally, we demonstrate the integration of multiomic information, where we found ceramide localization to be correlated with SMPD3 abundance (ceramide synthesis protein), and we also utilized protein abundance to resolve metabolite isomeric ambiguity. Overall, the integration of DESI-MSI into the multiomic workflow allows for complementary spatial- and molecular-level information to be achieved from optimized implementations of each MS assay inherent to the workflow itself.
pubs.acs.org
Reposted by Chris Anderton ⚜️
Kevin Zemaitis
@kevinzemaitis.bsky.social
· Dec 21

Post Doctorate RA - Spatial Glycomics Scientist in Richland, Washington | Pacific Northwest National Laboratory
PNNL is hiring a Post Doctorate RA - Spatial Glycomics Scientist in Richland, Washington. Review all of the job details and apply today!
careers.pnnl.gov
Reposted by Chris Anderton ⚜️
Chris Anderton ⚜️
@anderton.bsky.social
· Oct 22










