


Check out CiliaBuilder for ChimeraX → cxtoolshed.rbvi.ucsf.edu/apps/chimera...
Build custom 3D cilia & centriole models (tips + primary cilia included).
Grab the models here: makerworld.com/en/@user_763...

Grab the models here: makerworld.com/en/@user_763...
Here is a look at how I’m using CiliaBuilder in @UCSFChimeraX to generate accurate 3D prints of centrioles and cilia. Perfect for teaching or lab displays!
Watch the process: youtu.be/IKnnp3ovptQ #CryoEM #CellBio #ChimeraX


Here is a look at how I’m using CiliaBuilder in @UCSFChimeraX to generate accurate 3D prints of centrioles and cilia. Perfect for teaching or lab displays!
Watch the process: youtu.be/IKnnp3ovptQ #CryoEM #CellBio #ChimeraX
Check it out!
#cilia
www.nature.com/articles/s41...

Check it out!
#cilia
www.nature.com/articles/s41...
www.youtube.com/playlist?lis...

www.youtube.com/playlist?lis...
www.youtube.com/watch?v=SJHo...
If your installation didn't work a few days ago, try again. I've fixed a few things, and let me know if it still doesn't work for you.

www.youtube.com/watch?v=SJHo...
If your installation didn't work a few days ago, try again. I've fixed a few things, and let me know if it still doesn't work for you.
Video tutorial coming after the holidays
Video tutorial coming after the holidays
Check out CiliaBuilder for ChimeraX → cxtoolshed.rbvi.ucsf.edu/apps/chimera...
Build custom 3D cilia & centriole models (tips + primary cilia included).



Check out CiliaBuilder for ChimeraX → cxtoolshed.rbvi.ucsf.edu/apps/chimera...
Build custom 3D cilia & centriole models (tips + primary cilia included).
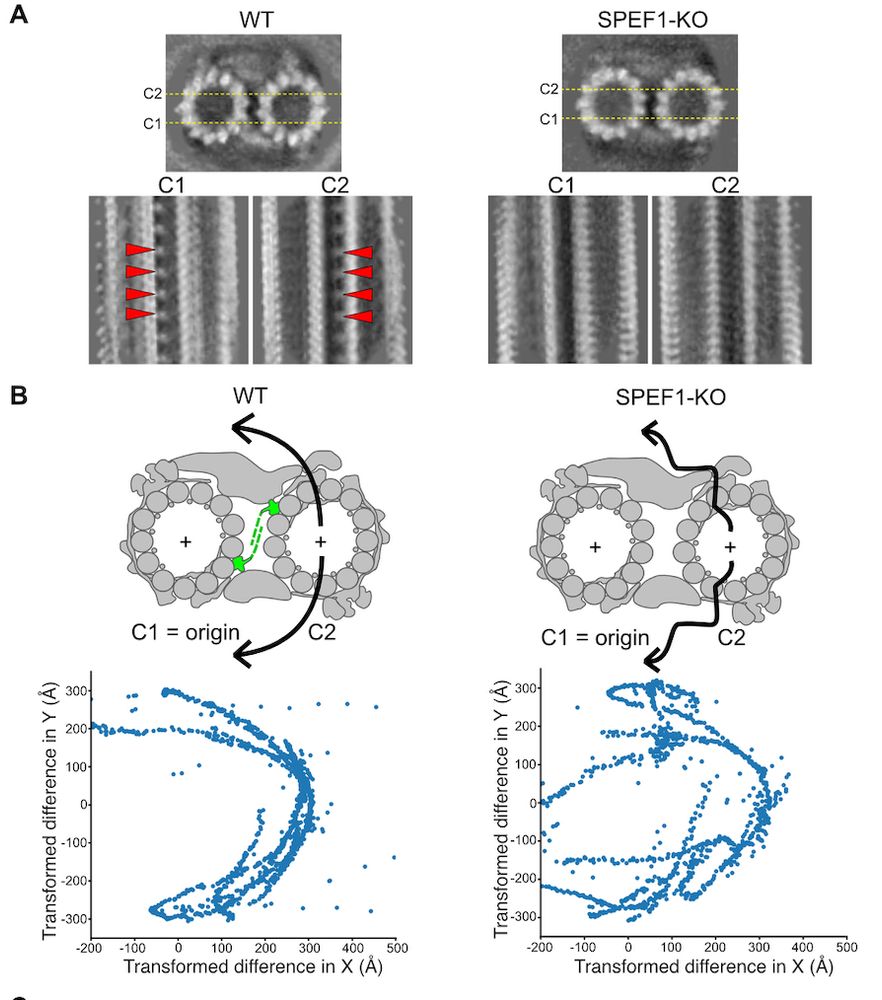
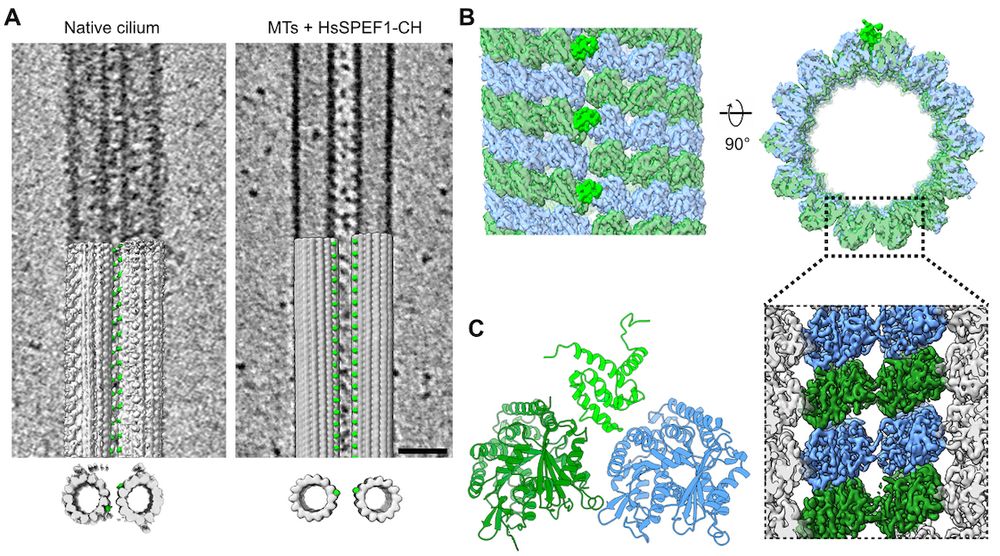
www.sciencedirect.com/science/arti...
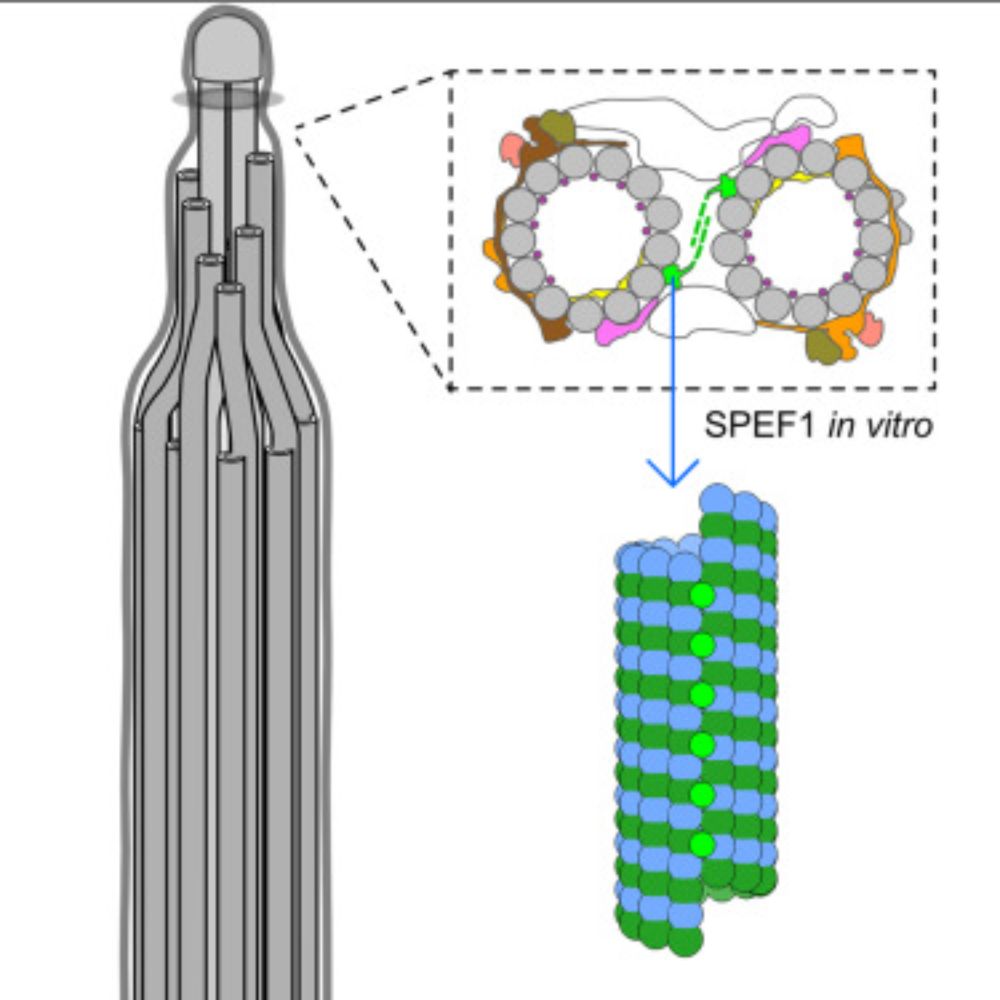
www.sciencedirect.com/science/arti...
@biorxiv-cellbio.bsky.social
www.biorxiv.org/content/10.1...

www.embopress.org/doi/full/10....
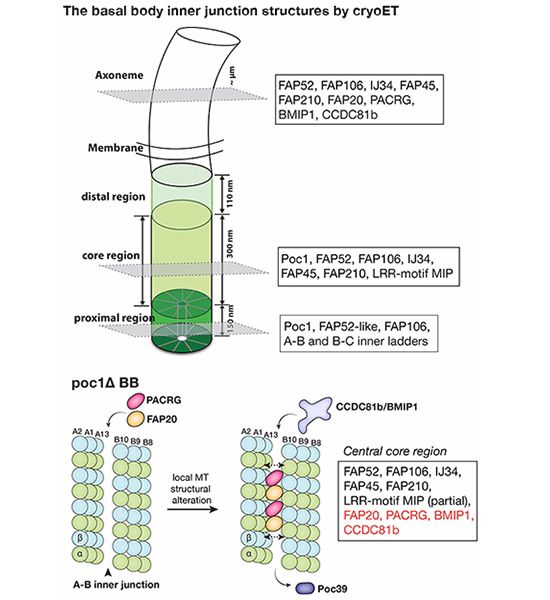
www.embopress.org/doi/full/10....
www.biorxiv.org/content/10.1...
1/8 🧵

www.biorxiv.org/content/10.1...
1/8 🧵
Check out the details here: www.biorxiv.org/content/10.1...

Check out the details here: www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
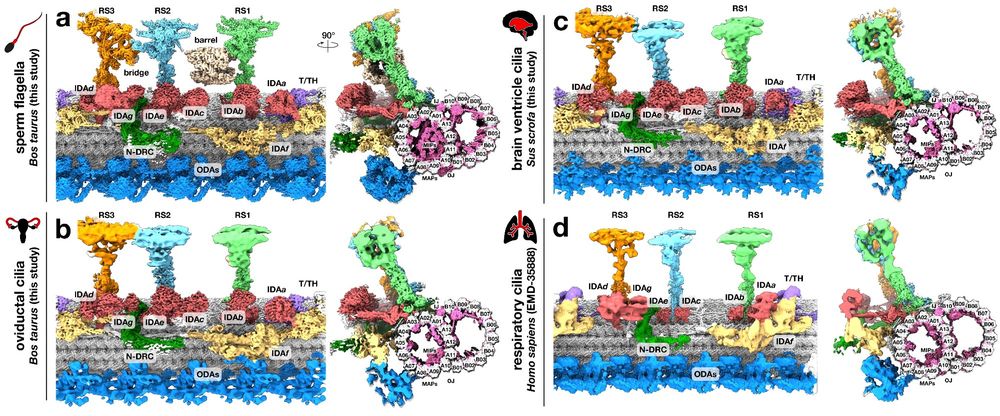
www.nature.com/articles/s41...

