Posts
Media
Videos
Starter Packs
builab.bsky.social
@builab.bsky.social
· Jul 11
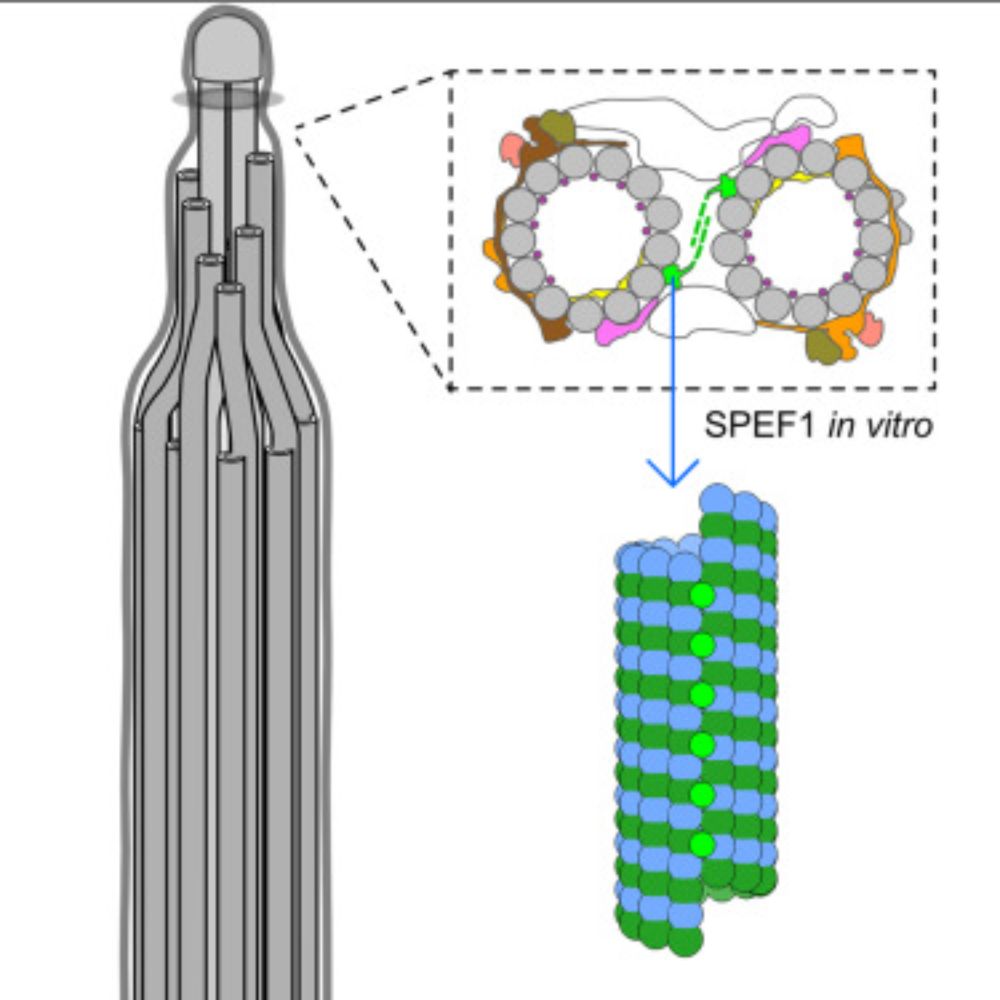
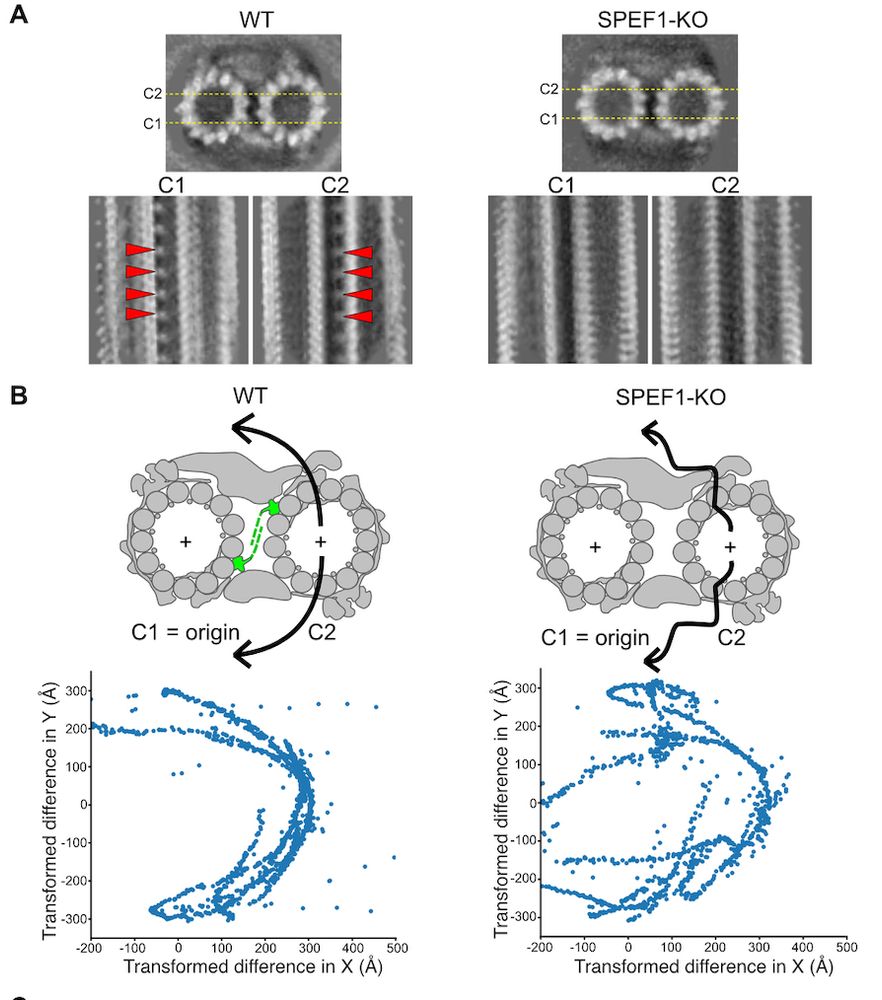
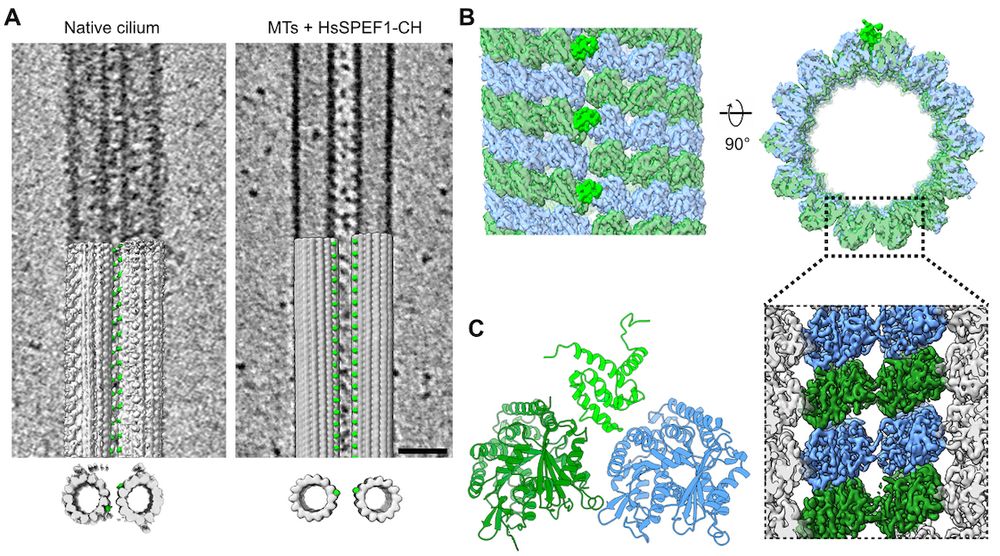
Structure of the ciliary tip central pair reveals the unique role of the microtubule-seam binding protein SPEF1
Motile cilia are unique organelles with the ability to move autonomously. The force generated by beating cilia propels cells and moves fluids. The cil…
www.sciencedirect.com
builab.bsky.social
@builab.bsky.social
· Feb 26
Reposted
Reposted
Yoshi Ichikawa
@ichikawa-lab.bsky.social
· Jan 28

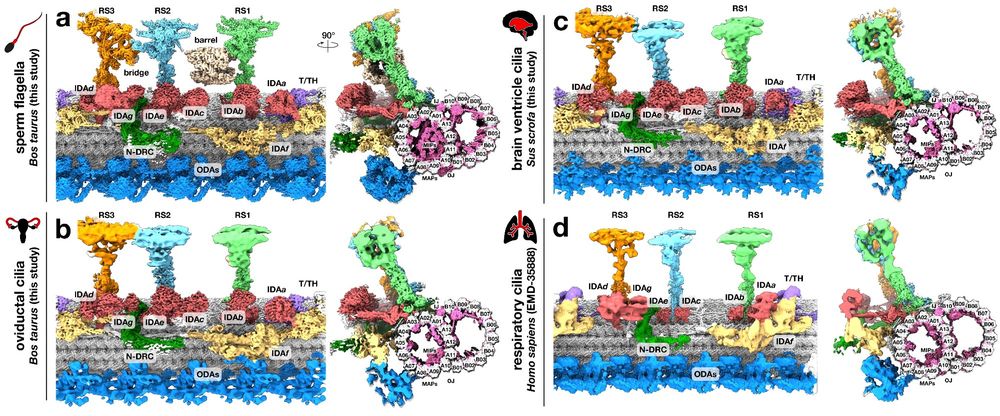
Dynein-2 is tuned for the A-tubule of the ciliary doublet through tubulin tyrosination
Eukaryotic cilia and flagella are thin structures present on the surface of cells, playing vital roles in signaling and cellular motion. Cilia structures rely on intraflagellar transport (IFT), which ...
www.biorxiv.org
Reposted
Pengxin Chai
@pengxinchai.bsky.social
· Jan 21

DNAHX: a novel, non-motile dynein heavy chain subfamily, identified by cryo-EM endogenously
Ciliogenesis and cilia motility rely on the coordinated actions of diverse dyneins, yet the complexity of these motor proteins in cilia has posed challenges for understanding their specific roles. Tra...
www.biorxiv.org
Reposted
builab.bsky.social
@builab.bsky.social
· Dec 17
Reposted
Reposted