Chang-Yu Chang 張昌祐
@changyuchang.bsky.social
62 followers
110 following
6 posts
Penn postdoc in biology | microbes, ecology, evolution | from Taiwan
Posts
Media
Videos
Starter Packs
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐
Jesse Shapiro
@bjesseshapiro.bsky.social
· Aug 21
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐
Alvaro Sanchez
@asanchezlab.bsky.social
· Jul 16
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐
Will Shoemaker
@shoestrapped.bsky.social
· May 28
Reposted by Chang-Yu Chang 張昌祐
Alvaro Sanchez
@asanchezlab.bsky.social
· May 27
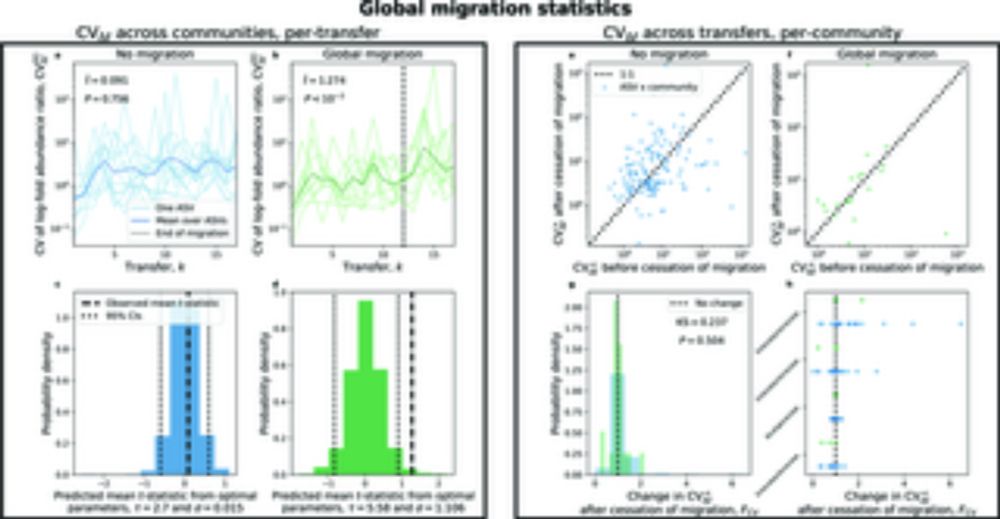
Macroecological patterns in experimental microbial communities
Author summary Determining whether an empirical pattern can be manipulated is a crucial step towards building a predictive theory. Our study aimed to determine the extent that experimental manipulatio...
journals.plos.org
Reposted by Chang-Yu Chang 張昌祐
Mike Blazanin
@mikeblazanin.bsky.social
· May 27

Quantifying phage infectivity from characteristics of bacterial population dynamics
A frequent goal of phage biology is to quantify how well a phage kills a population of host bacteria. Unfortunately, traditional methods to quantify phage success can be time-consuming, limiting the t...
doi.org
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐
Dmitri Petrov
@petrovadmitri.bsky.social
· May 15
Reposted by Chang-Yu Chang 張昌祐
SeppeKuehnLab
@seppekuehnlab.bsky.social
· Mar 31

Collective Microbial Effects Drive Toxin Bioremediation and Enable Rational Design
The metabolic activity of microbial communities is essential for host and environmental health, influencing processes from immune regulation to bioremediation. Given this importance, the rational desi...
www.biorxiv.org
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐
Reposted by Chang-Yu Chang 張昌祐