Website: novoalab.com
Big thanks to James for presenting his work on advanced CRISPR tools for epitranscriptomic research, and to @eu-life.bsky.social for this opportunity!🧬✈️
#EULIFE #CRG

Big thanks to James for presenting his work on advanced CRISPR tools for epitranscriptomic research, and to @eu-life.bsky.social for this opportunity!🧬✈️
#EULIFE #CRG
Congrats to @soniacruciani.bsky.social (former Novoa Lab PhD, now postdoc at the Deplancke Lab, EPFL) and @evamarianovoa.bsky.social👏
📖 www.nature.com/articles/s41...
Congrats to @soniacruciani.bsky.social (former Novoa Lab PhD, now postdoc at the Deplancke Lab, EPFL) and @evamarianovoa.bsky.social👏
📖 www.nature.com/articles/s41...
@novoalab.bsky.social !😀📌 Do you want to barcode up to 96 #RNA samples in your #nanopore flowcells? How well do RNA #modification-aware models perform? What if there is #noBasecallingModel for your modification-of-interest? Find it out here!😊 www.biorxiv.org/content/10.1...

@novoalab.bsky.social !😀📌 Do you want to barcode up to 96 #RNA samples in your #nanopore flowcells? How well do RNA #modification-aware models perform? What if there is #noBasecallingModel for your modification-of-interest? Find it out here!😊 www.biorxiv.org/content/10.1...
We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)

We systematically benchmarked @nanoporetech.com 's modification-aware basecalling models released for RNA on sets of in vitro and in vivo sequences and made some curious observations 🧬🔍.
bit.ly/4lXqNul
Follow along for a little recap (1/12)



t.co/IAld0ocjDf
@nickkouvelas.bsky.social @unibern.bsky.social #biorxiv #mRNA #Ribosome

t.co/IAld0ocjDf
@nickkouvelas.bsky.social @unibern.bsky.social #biorxiv #mRNA #Ribosome
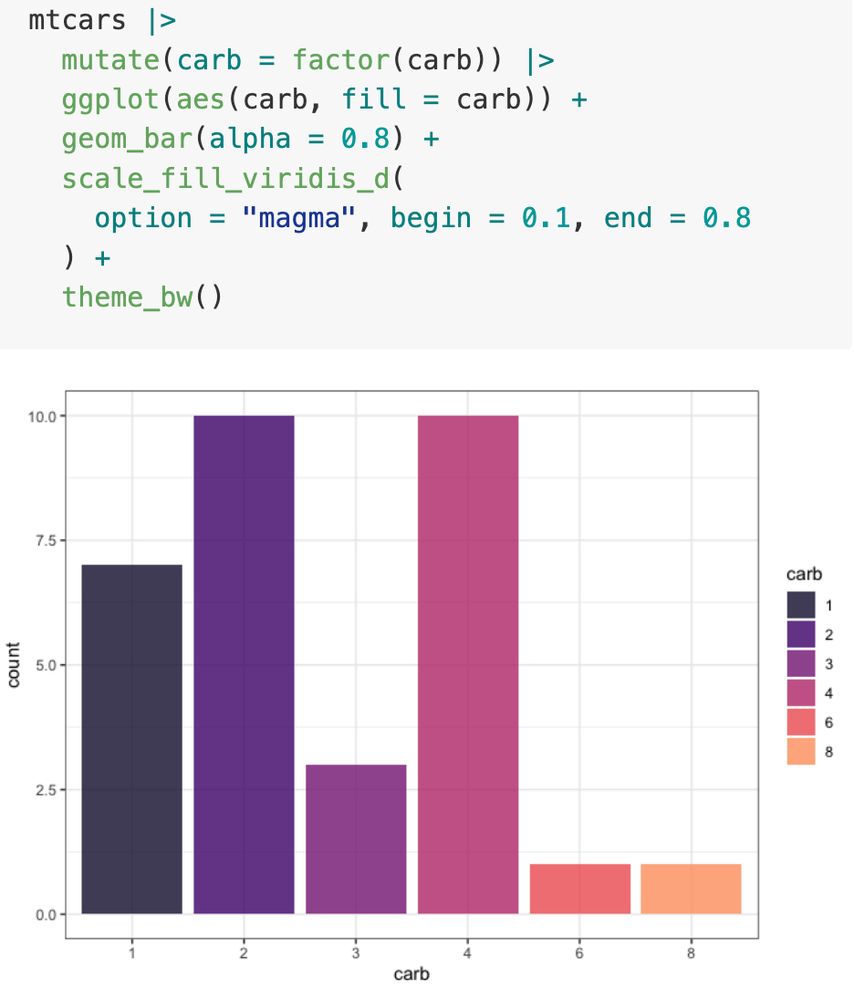
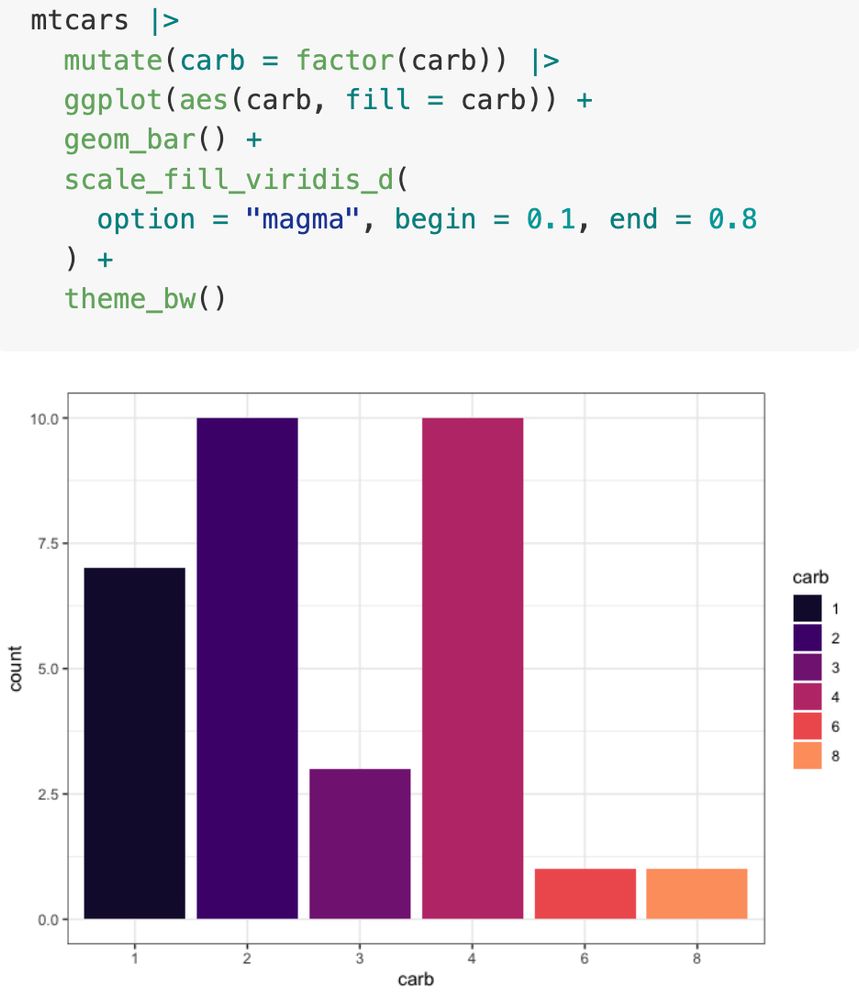
Compare left versus right.
Compare left versus right.




