


Sex and smoking bias in the selection of somatic mutations in human bladder
www.nature.com/articles/s41...
by @raquelbmi.bsky.social, @ferriol.bsky.social et al (in collaboration with Rosana Risques lab in @uwmedicine.bsky.social)

Sex and smoking bias in the selection of somatic mutations in human bladder
www.nature.com/articles/s41...
by @raquelbmi.bsky.social, @ferriol.bsky.social et al (in collaboration with Rosana Risques lab in @uwmedicine.bsky.social)

(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...

(1) Accurate sequencing of sperm at scale
(2) Positive selection of spermatogenesis driver mutations across the exome
(3) Offspring disease risks from male reproductive aging
[1/n]
www.nature.com/articles/s41...
brnw.ch/21wRUZZ

brnw.ch/21wRUZZ

www.nature.com/articles/s41...
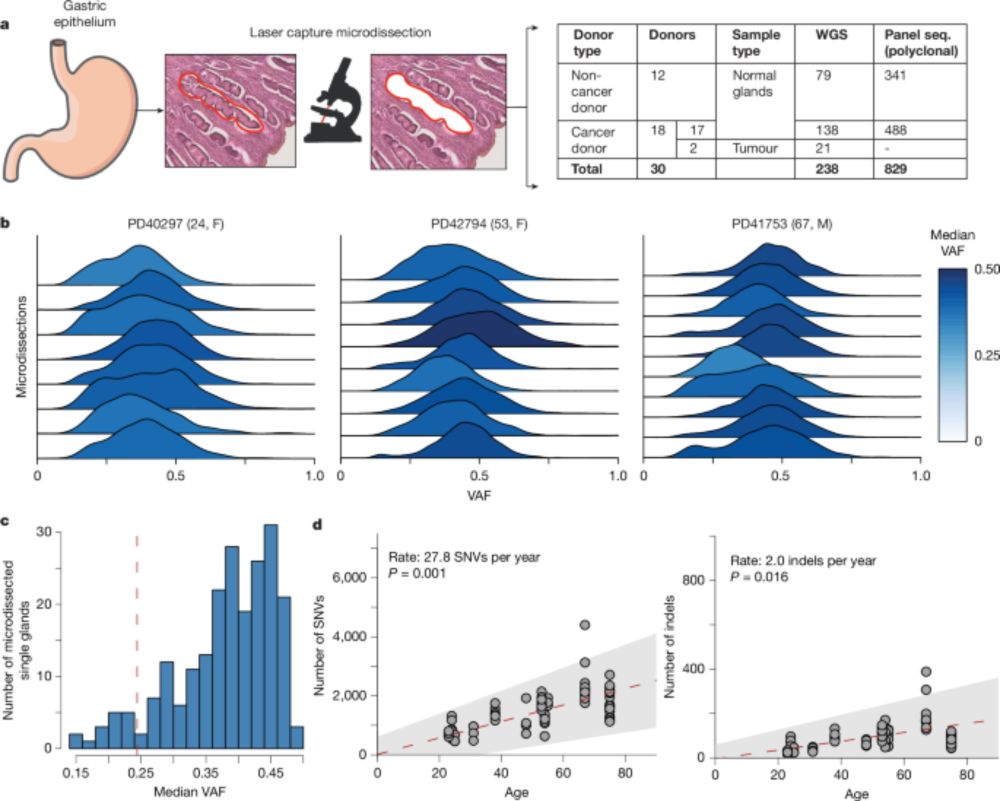
www.nature.com/articles/s41...
www.nature.com/articles/s41...
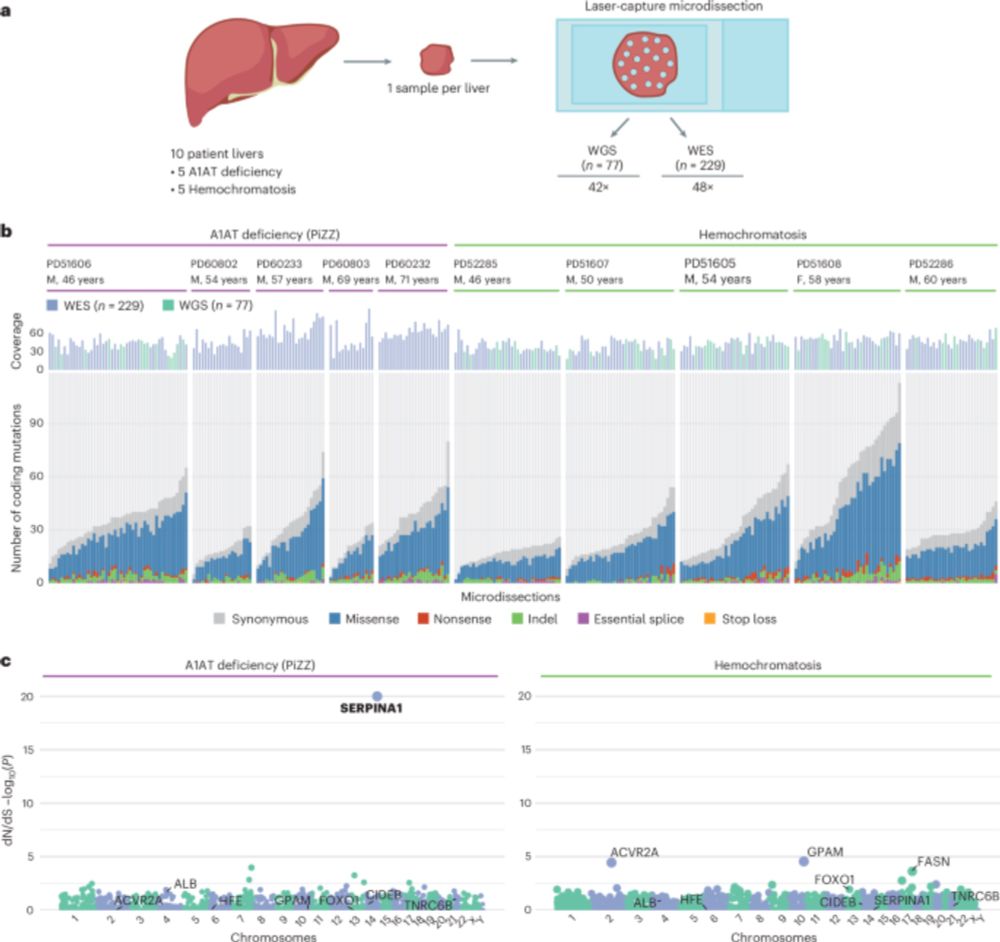
www.nature.com/articles/s41...
Takehome: Widespread and substantial deceleration in fitness with age!
Amazing effort by PhD student Hamish MacGregor 💪
www.biorxiv.org/content/10.1...
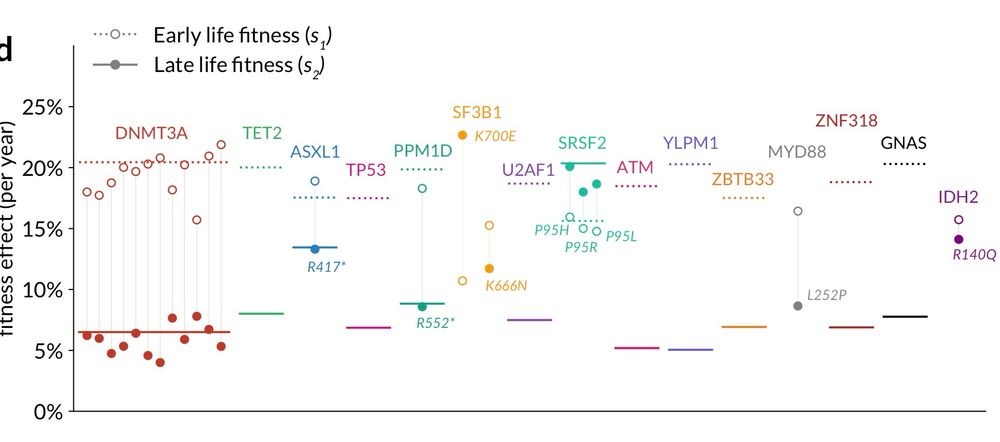
Takehome: Widespread and substantial deceleration in fitness with age!
Amazing effort by PhD student Hamish MacGregor 💪
www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
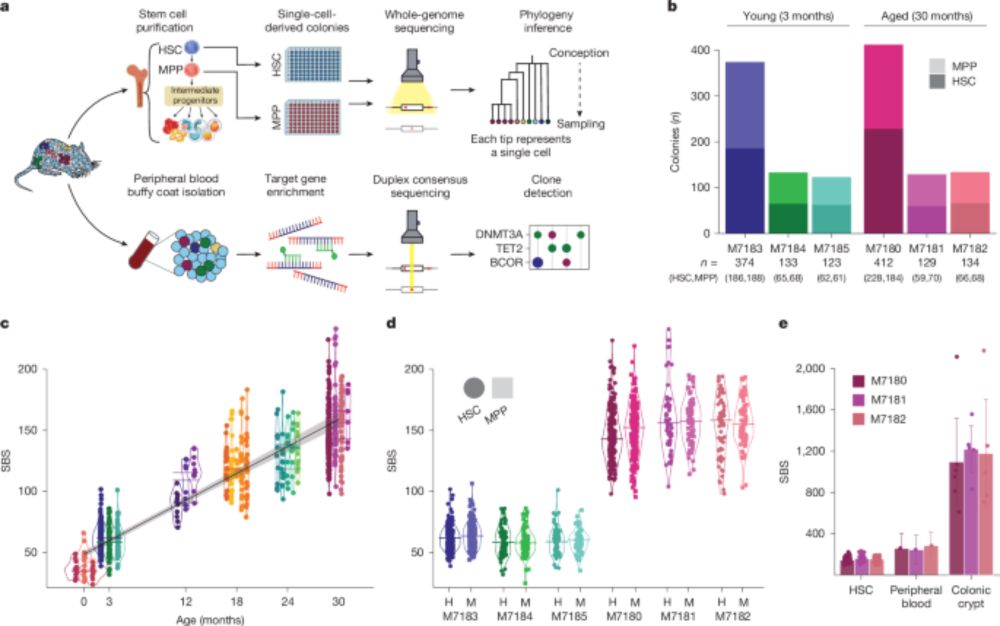
www.nature.com/articles/s41...
For me this was discovery science as I had always hoped it would be. A lot of fun, and some proper detective work with plenty of twists & turns on the way. Brief thread below

For me this was discovery science as I had always hoped it would be. A lot of fun, and some proper detective work with plenty of twists & turns on the way. Brief thread below

www.nature.com/articles/s41...
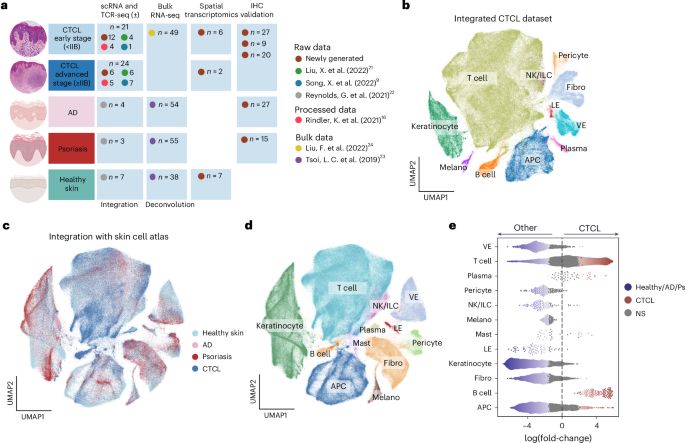
www.nature.com/articles/s41...


