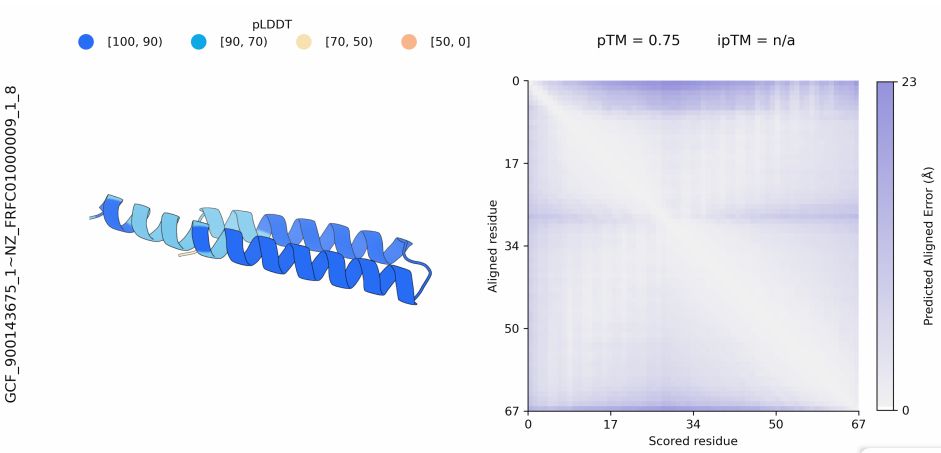
Interested in hydrogenases, evolution, protein design.
💻 https://www.jameslingford.com/
www.statnews.com/2024/06/20/r...

www.statnews.com/2024/06/20/r...





📑 Paper doi.org/10.1038/s443...
🔭 Explore virosphere viro3d.cvr.gla.ac.uk

📑 Paper doi.org/10.1038/s443...
🔭 Explore virosphere viro3d.cvr.gla.ac.uk
evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations

evolutionary history journals.asm.org/doi/10.1128/...
A review on structural phylogenetics utility and limitations
academic.oup.com/gbe/article/...

academic.oup.com/gbe/article/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
github.com/RosettaCommo...
You can now design proteins, and in particular enzymes from just partially defined amino acid side chains, and without defining their sequence position or order!

github.com/RosettaCommo...
You can now design proteins, and in particular enzymes from just partially defined amino acid side chains, and without defining their sequence position or order!
www.science.org/doi/10.1126/...
github.com/lehner-lab/c...

www.science.org/doi/10.1126/...
github.com/lehner-lab/c...
• it's a bad idea to install a tool on your machine that has vast read+write permissions
• code that works but is really outputting false info is a real danger and hard to catch. LLMs make that danger worse
• it's a bad idea to install a tool on your machine that has vast read+write permissions
• code that works but is really outputting false info is a real danger and hard to catch. LLMs make that danger worse
www.sciencedirect.com/science/arti... From @hkws.bsky.social

www.sciencedirect.com/science/arti... From @hkws.bsky.social
- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍

- SIMD FW/BW alignment (preprint soon!)
- Sub. Mat. λ calculator by Eric Dawson
- Faster ARM SW by Alexander Nesterovskiy
- MSA-Pairformer’s proximity-based pairing for multimer prediction (www.biorxiv.org/content/10.1...; avail. in ColabFold API)
💾 github.com/soedinglab/M... & 🐍
github.com/chevrettelab...
github.com/chevrettelab...
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
We tested it on the GTDB r220 archaeal supermatrix (5,869 taxa & 10,101 cols) removing 55% of sites in <2h.
The phylogeny showed several interesting groupings with overall improved branch support:
#phylogenetics #ArchaeaSky #MSA #opensource #MEvoSky #MicroSky

We tested it on the GTDB r220 archaeal supermatrix (5,869 taxa & 10,101 cols) removing 55% of sites in <2h.
The phylogeny showed several interesting groupings with overall improved branch support:
#phylogenetics #ArchaeaSky #MSA #opensource #MEvoSky #MicroSky
tgv download hg38
Download UCSC reference genomes to a local sqlite db for much faster browsing. Awesome Rust tools (twobit, bigtools) made this simple.
github.com/zeqianli/tgv
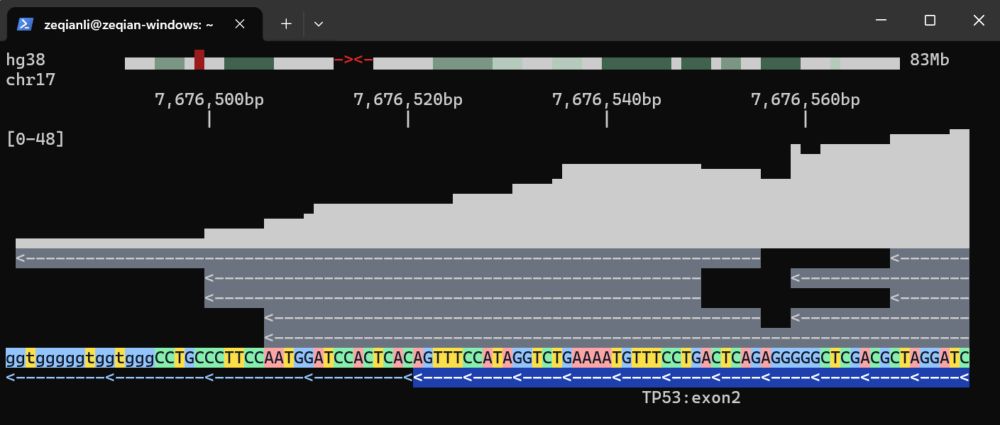
tgv download hg38
Download UCSC reference genomes to a local sqlite db for much faster browsing. Awesome Rust tools (twobit, bigtools) made this simple.
github.com/zeqianli/tgv

Never have to abandon my precious vim setup again
Never have to abandon my precious vim setup again
www.nature.com/articles/s41...
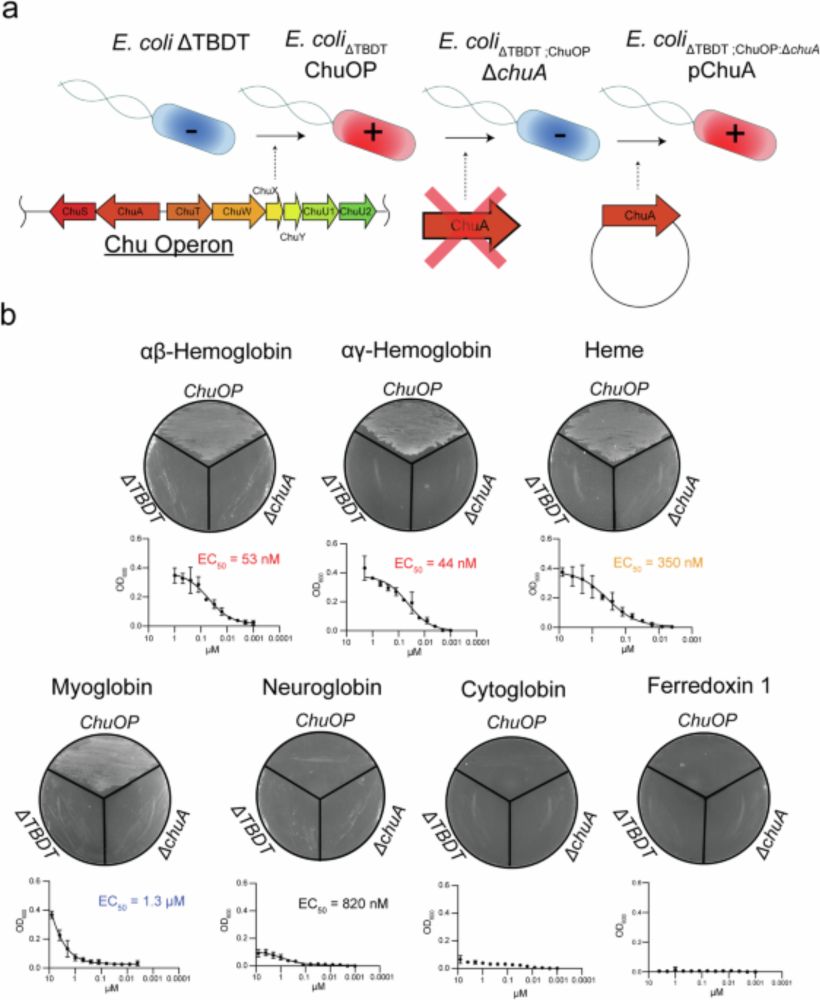
www.nature.com/articles/s41...
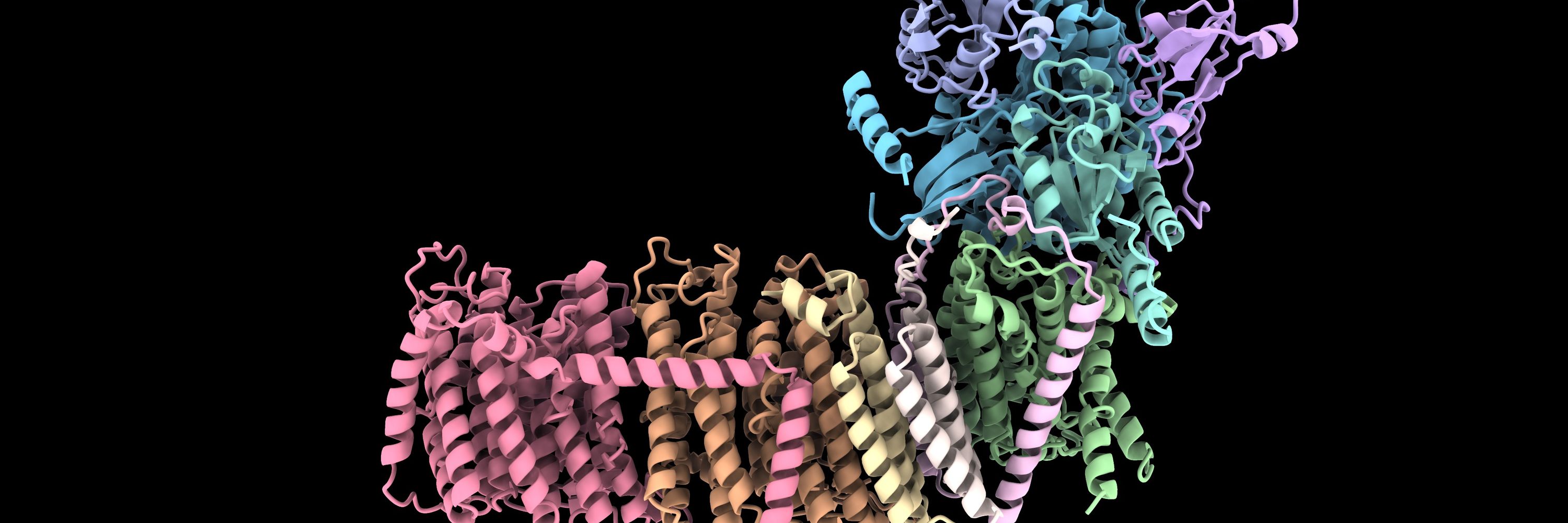
📄 www.biorxiv.org/content/10.1...
🌐 search.foldseek.com/folddisco
github.com/martinpacesa...
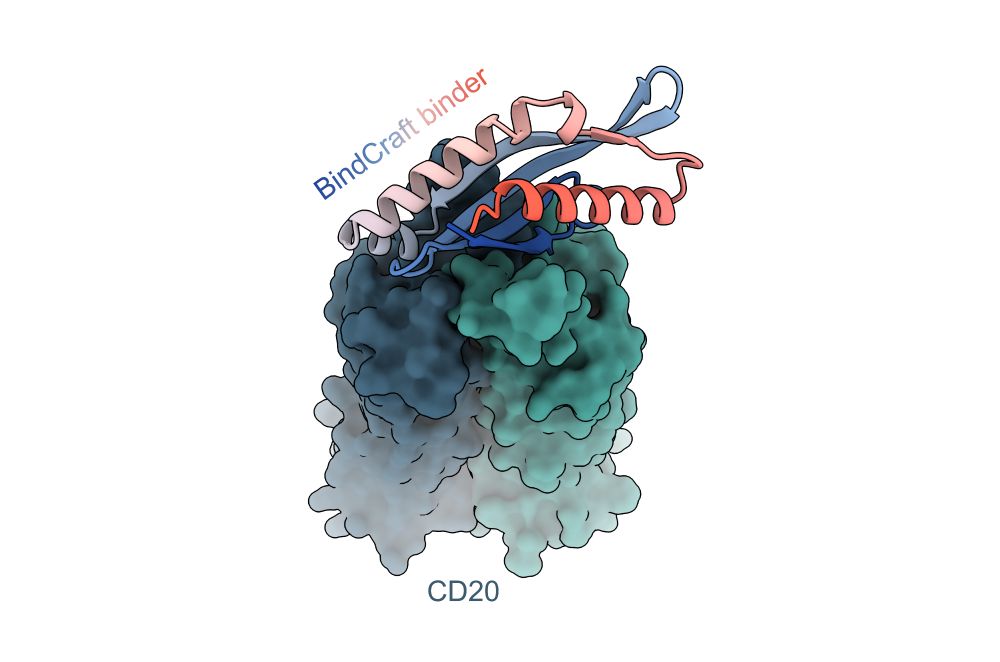
github.com/martinpacesa...