genetics-gsa.org/yeast-2026/

genetics-gsa.org/yeast-2026/
#PhD #PhDSky

#PhD #PhDSky
We report the existence of a previously uncharacterized signaling pathway that is responsible for activating cell death upon loss of gene expression.
1/n 🧪
www.cell.com/cell/fulltex...

We report the existence of a previously uncharacterized signaling pathway that is responsible for activating cell death upon loss of gene expression.
1/n 🧪
www.cell.com/cell/fulltex...
Read more: buff.ly/AhEqx0f

Read more: buff.ly/AhEqx0f
@fbm-unil.bsky.social @unil.bsky.social @umasschan.bsky.social, on the conservation of genetic suppression interactions across genetic contexts, now published in G3 @genetics-gsa.bsky.social: doi.org/10.1093/g3jo...
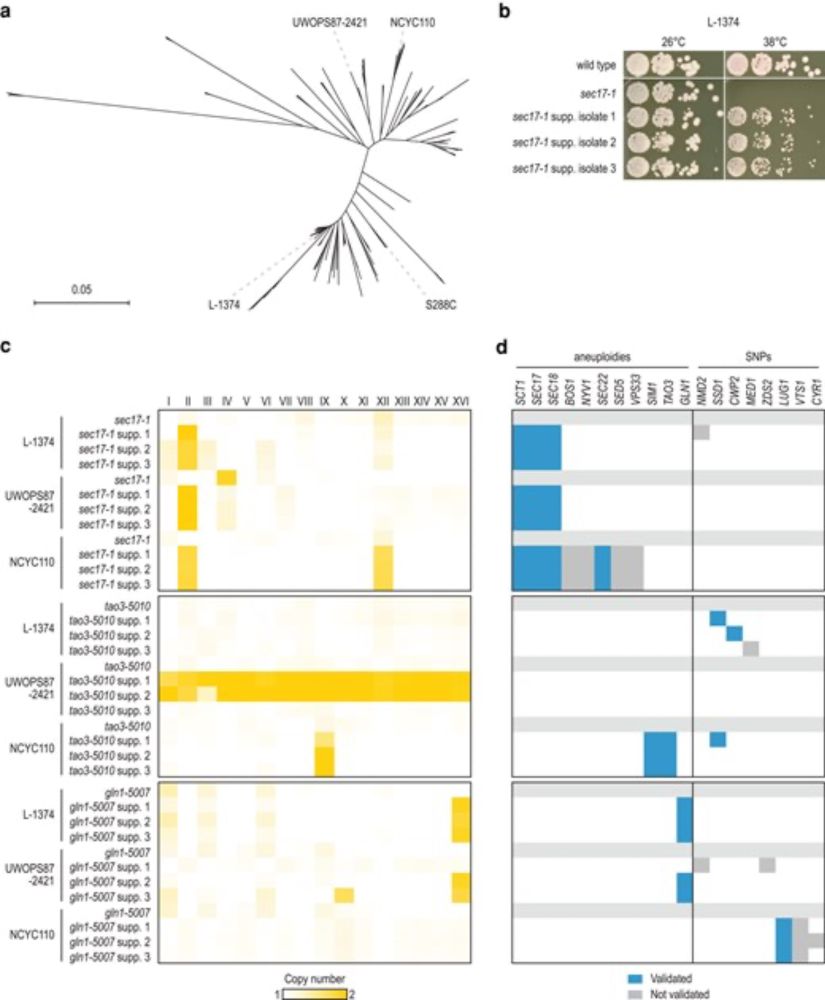
@fbm-unil.bsky.social @unil.bsky.social @umasschan.bsky.social, on the conservation of genetic suppression interactions across genetic contexts, now published in G3 @genetics-gsa.bsky.social: doi.org/10.1093/g3jo...
So what did we find?

So what did we find?
www.science.org/doi/10.1126/...
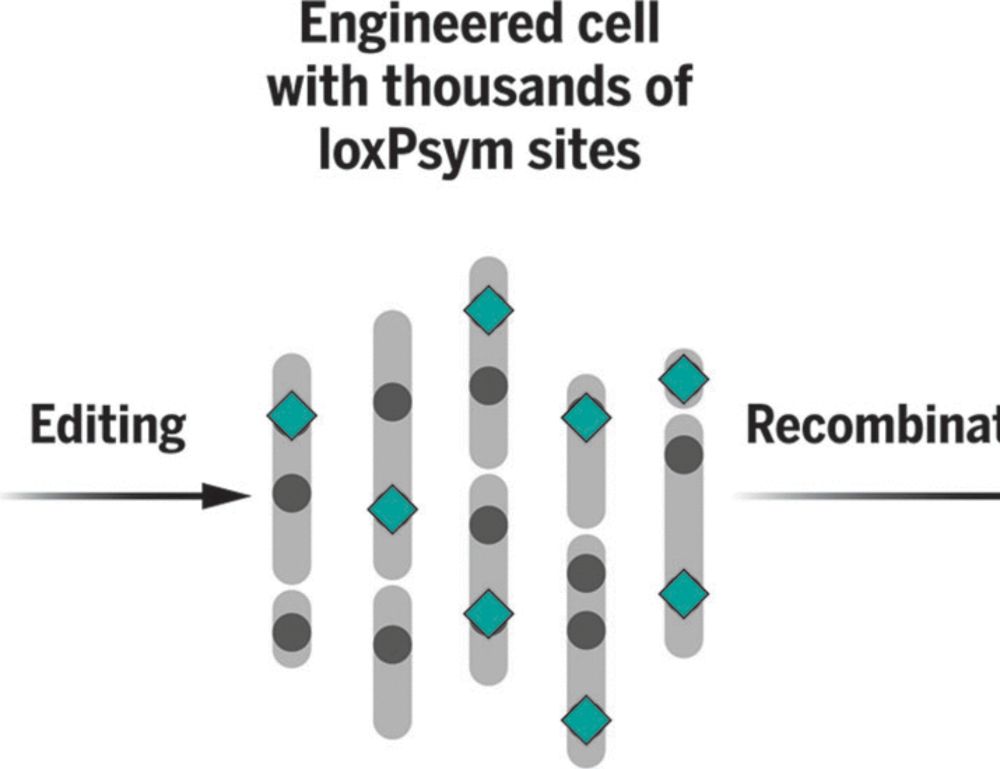
www.science.org/doi/10.1126/...
July 22 - August 12, 2025
Application Deadline: March 31, 2025
Arrival: July 22nd by 6pm EST
Departure: August 12th around 12pm EST
For more info and to apply please visit the Yeast Genetics & Genomics course website: meetings.cshl.edu/courses.aspx...

July 22 - August 12, 2025
Application Deadline: March 31, 2025
Arrival: July 22nd by 6pm EST
Departure: August 12th around 12pm EST
For more info and to apply please visit the Yeast Genetics & Genomics course website: meetings.cshl.edu/courses.aspx...
Registration and abstract submission are NOW OPEN!
Join us at the 32nd ICYGMB
📅 Save the Date: 21-24 July 2025
📍 Location: Paris @sorbonne-universite.fr
🔗 Submit: premc.org/yeast2025/su...
🕒 Register now to take advantage of the Early Bird fee premc.org/yeast2025/re...

Registration and abstract submission are NOW OPEN!
Join us at the 32nd ICYGMB
📅 Save the Date: 21-24 July 2025
📍 Location: Paris @sorbonne-universite.fr
🔗 Submit: premc.org/yeast2025/su...
🕒 Register now to take advantage of the Early Bird fee premc.org/yeast2025/re...
bit.ly/40cIfSl
@fritzroth.bsky.social @jsvanleeuwen.bsky.social @pedrobeltrao.bsky.social

meetings.cshl.edu/meetings.asp...

meetings.cshl.edu/meetings.asp...
We have Assistant and Associate Professor positions! 🚀
Pittsburgh is beautiful, and we are doing amazing science here.
cfopitt.taleo.net/careersectio...
cfopitt.taleo.net/careersectio...
Please apply by Dec 2!
We have Assistant and Associate Professor positions! 🚀
Pittsburgh is beautiful, and we are doing amazing science here.
cfopitt.taleo.net/careersectio...
cfopitt.taleo.net/careersectio...
Please apply by Dec 2!

