Statistical genomics. Currently working on single-cell data analysis in context of drug development. Previously post-doc at UC Berkeley and Ghent University.

You will be embedded in our Discovery Statistics organization, and supported by myself and colleagues. Feel free to reach out for any additional information.
tinyurl.com/36uyca7f
Reposted by Cyrus Samii, Koen Van Den Berge
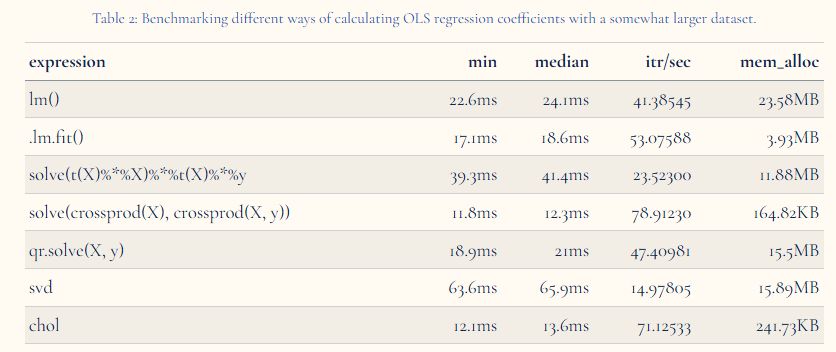
In this post, I explain different approaches for solving linear regression in R: directly, using QR, singular value and Cholesky decompositions, and do some benchmarking for comparison with in-built approaches.
thomvolker.github.io/blog/2506_re...
Reposted by Koen Van Den Berge

I'm so excited about this!
www.biorxiv.org/content/10.1...
Reposted by Koen Van Den Berge

www.biorxiv.org/content/10.1...
Reposted by Koen Van Den Berge, Andy Reisinger

Reposted by Daniel Vaulot, Koen Van Den Berge
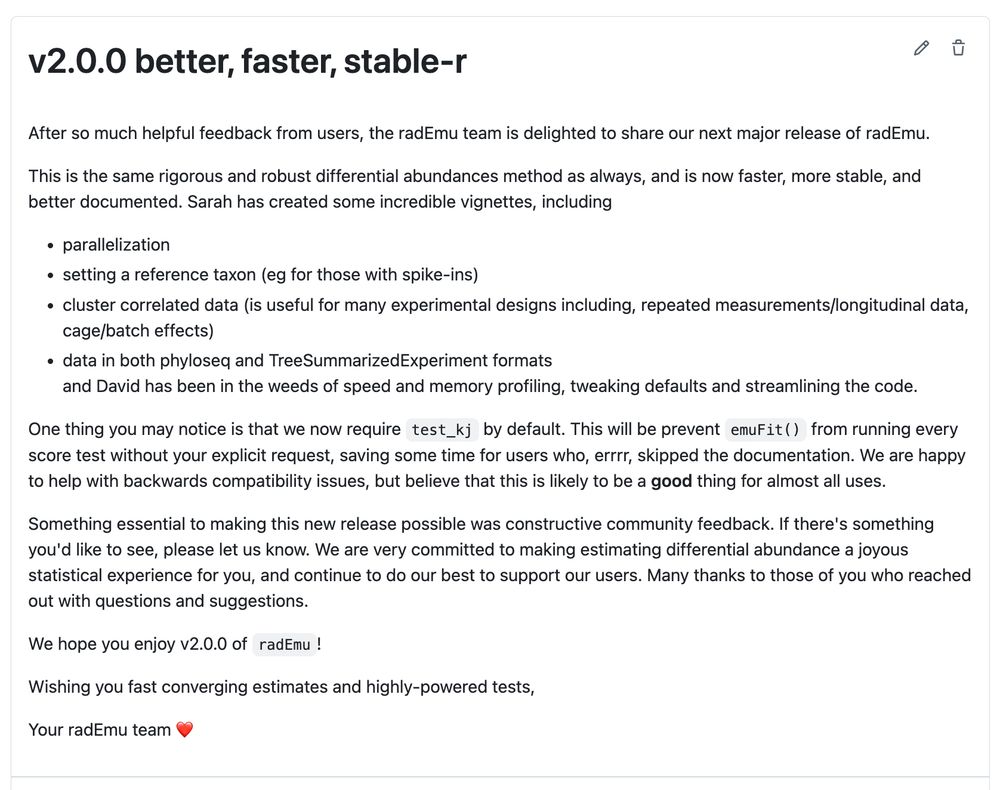
`remotes::install_github("statdivlab/radEmu")`
A huge thanks to users for sharing their requests and questions, and to the maintenance team (Sarah and @davidandacat.bsky.social ) for their time and commitment!
Release notes: github.com/statdivlab/r...
Reposted by Koen Van Den Berge
Reposted by Koen Van Den Berge

Sparser and more interpretable than the lasso. We're excited! arxiv.org/abs/2501.18360
R: github.com/trevorhastie...
Reposted by Koen Van Den Berge
arxiv.org/abs/2412.20509
We present a stochastic gradient descent method that allows to efficiently and very quickly estimate latent factors for, e.g., dimensionality reduction of single-cell data
Reposted by Koen Van Den Berge
www.encodeproject.org/single-cell/...
Reposted by Koen Van Den Berge
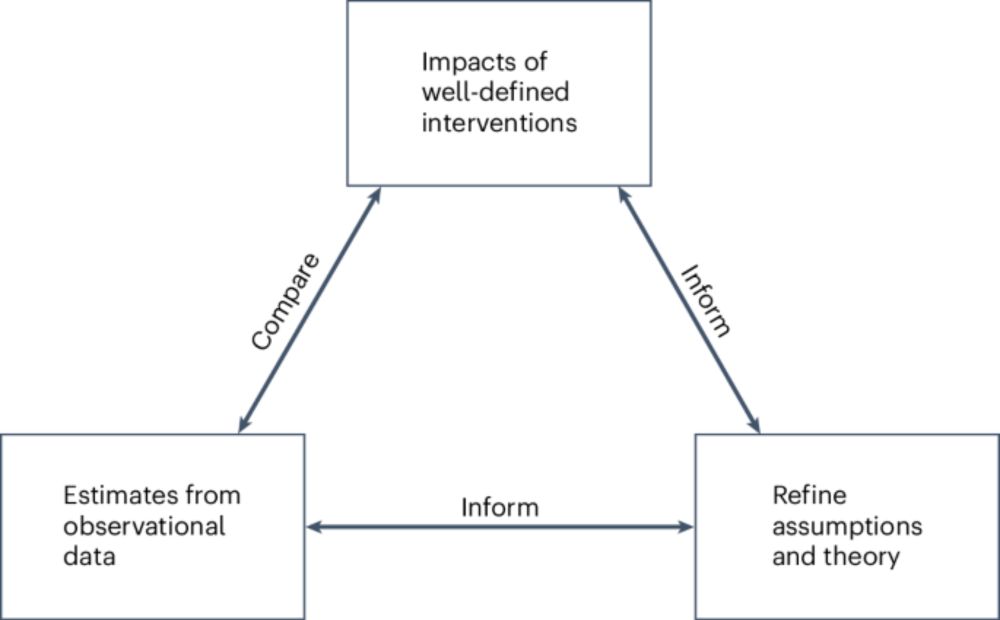
Causal inference is hard:
www.nature.com/articles/s41...
Reposted by Koen Van Den Berge

Luckily for you, @aaronkwc.bsky.social has!
Aaron will help you grok:
What's going on?
What is TF-IDF?
Is there really single-cell level chromatin information?
Check it out 👇
www.biorxiv.org/content/10.1...
🧪🧬💻

