
Lab Website: forsberglab.org
Banner Art: Tamanash Bhattacharya
In this work, we use functional metagenomics to find phage defenses from human and soil microbiomes!
Congrats to first author @luis840alberto.bsky.social !
www.cell.com/cell-host-mi...
(1/3)
I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡
Continue reading (🧵)

I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡
Continue reading (🧵)
Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8

Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8
blog.genesmindsmachines.com/p/we-still-c...

blog.genesmindsmachines.com/p/we-still-c...
Menstruation is understudied due to societal taboos + a biological challenge: mice (a key system for research + drug discovery) don’t menstruate.
@cagricevrim.bsky.social made menstruating mice + used them to discover early events in menstruation.
He is on the job market!
@karalmckinley.bsky.social
We built the first transgenic model of menstruation in mice.
We used it to uncover how the endometrium organizes and sheds during menstruation. 🧪
www.biorxiv.org/content/10.1...
🧵

Menstruation is understudied due to societal taboos + a biological challenge: mice (a key system for research + drug discovery) don’t menstruate.
@cagricevrim.bsky.social made menstruating mice + used them to discover early events in menstruation.
He is on the job market!
~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...

~27,000 predicted viral protein monomers & homodimers
Conserved folds across bacteria, archaea & eukaryotic viruses
New toxin–antitoxin system KreTA uncovered
Vast “functional darkness” remains uncharted
www.science.org/doi/10.1126/...

(also, I love the name 'kleptosome' for an organelle).
A host organelle (in sea slugs) integrates stolen chloroplasts for animal photosynthesis

(also, I love the name 'kleptosome' for an organelle).
I've been very happy at UTSW Micro. Why?
Its a supportive environment full of excellent people. There's strong ambition, reflexive empathy, and excellent resources.

I've been very happy at UTSW Micro. Why?
Its a supportive environment full of excellent people. There's strong ambition, reflexive empathy, and excellent resources.
It was a bit tricky to figure out how to make everybody happy (you could put that on my epitaph...)
A🧵 on how I threaded this needle of compliance!
Stay tuned for summary thread on that experience…
(3/3)
It was a bit tricky to figure out how to make everybody happy (you could put that on my epitaph...)
A🧵 on how I threaded this needle of compliance!
In this work, we use functional metagenomics to find phage defenses from human and soil microbiomes!
Congrats to first author @luis840alberto.bsky.social !
www.cell.com/cell-host-mi...
(1/3)
In this work, we use functional metagenomics to find phage defenses from human and soil microbiomes!
Congrats to first author @luis840alberto.bsky.social !
www.cell.com/cell-host-mi...
(1/3)
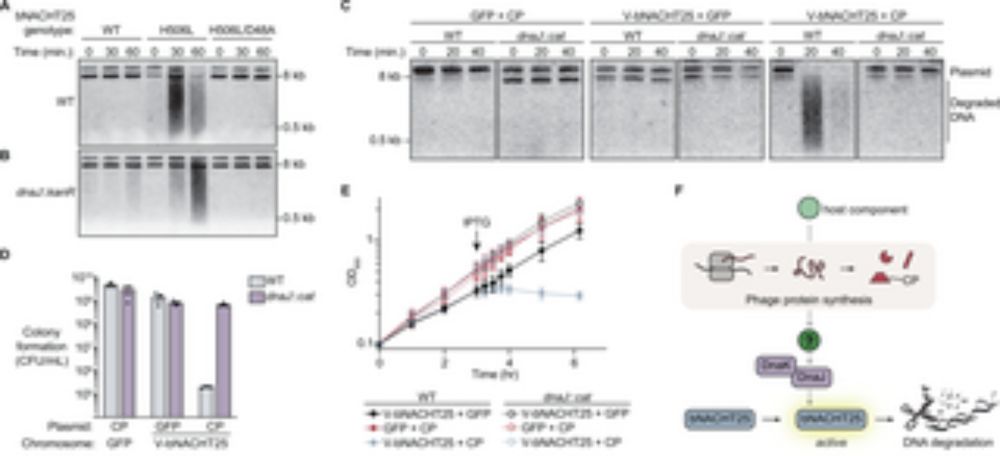
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
Full story here:
www.biorxiv.org/content/10.1...
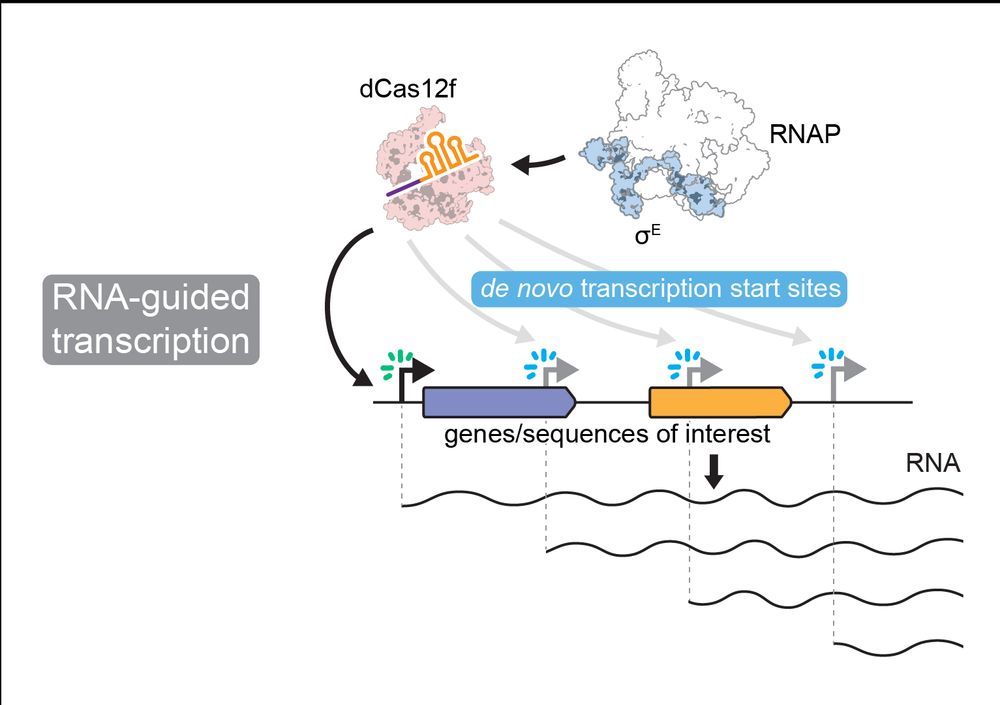
Full story here:
www.biorxiv.org/content/10.1...
plos.io/4koXP60
We used functional metagenomics to identify phage defenses in human and soil microbiomes. We scaled these selections while maintaining accuracy, enabling us to examine 9 habitats for defense elements against 7 phages.
www.biorxiv.org/content/10.1...
1/10
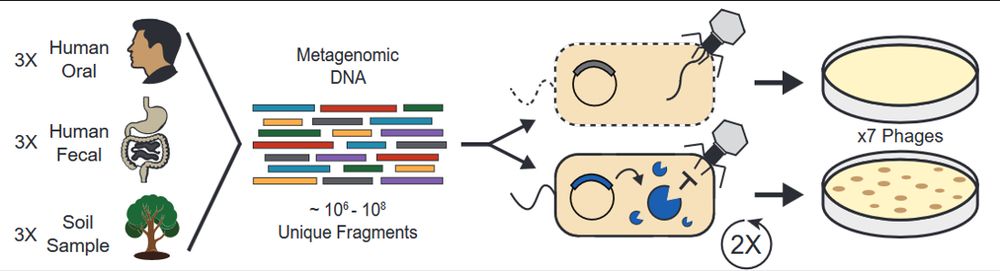
We used functional metagenomics to identify phage defenses in human and soil microbiomes. We scaled these selections while maintaining accuracy, enabling us to examine 9 habitats for defense elements against 7 phages.
www.biorxiv.org/content/10.1...
1/10
1. The importance of stupidity in scientific research
Open Access
journals.biologists.com/jcs/article/...

1. The importance of stupidity in scientific research
Open Access
journals.biologists.com/jcs/article/...
Not so "systematic" as very few known, fun stuff on MGE, antidefense islands and cool phages exaptations!
academic.oup.com/nar/advance-...
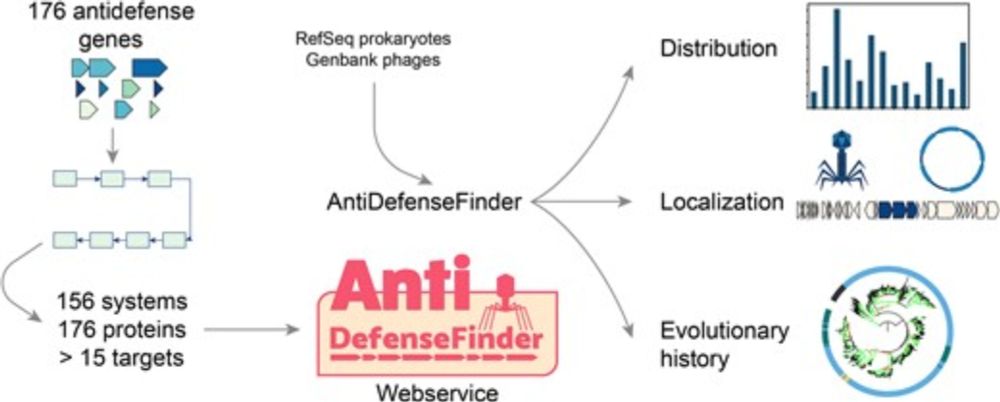
Not so "systematic" as very few known, fun stuff on MGE, antidefense islands and cool phages exaptations!
academic.oup.com/nar/advance-...
Please help me amplify a special faculty search. We're looking for someone in "Microbial Systems Biology" to join me and @kreynoldslab.bsky.social at UTSW Sys Bio / Bioinformatics.
There is an empty lab space right next to us waiting for you!
apply.interfolio.com/154339
1/4


Please help me amplify a special faculty search. We're looking for someone in "Microbial Systems Biology" to join me and @kreynoldslab.bsky.social at UTSW Sys Bio / Bioinformatics.
There is an empty lab space right next to us waiting for you!
apply.interfolio.com/154339
1/4
I've been very, very happy at UTSW Micro.
Supportive environment. Strong ambition. Strong empathy. Excellent resources.
jobs.utsouthwestern.edu/job/20847915...

I've been very, very happy at UTSW Micro.
Supportive environment. Strong ambition. Strong empathy. Excellent resources.
jobs.utsouthwestern.edu/job/20847915...
Microbiology & Molecular Genetics at University of Pittsburgh School of Medicine invites applicants for tenure-track microbiology faculty at the Assistant or Associate Professor level. Our scope is broad, we seek creative colleagues! More info: tinyurl.com/PittMMG-Jobs

Microbiology & Molecular Genetics at University of Pittsburgh School of Medicine invites applicants for tenure-track microbiology faculty at the Assistant or Associate Professor level. Our scope is broad, we seek creative colleagues! More info: tinyurl.com/PittMMG-Jobs
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

