🌐 https://emmanuelczt.github.io/

Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
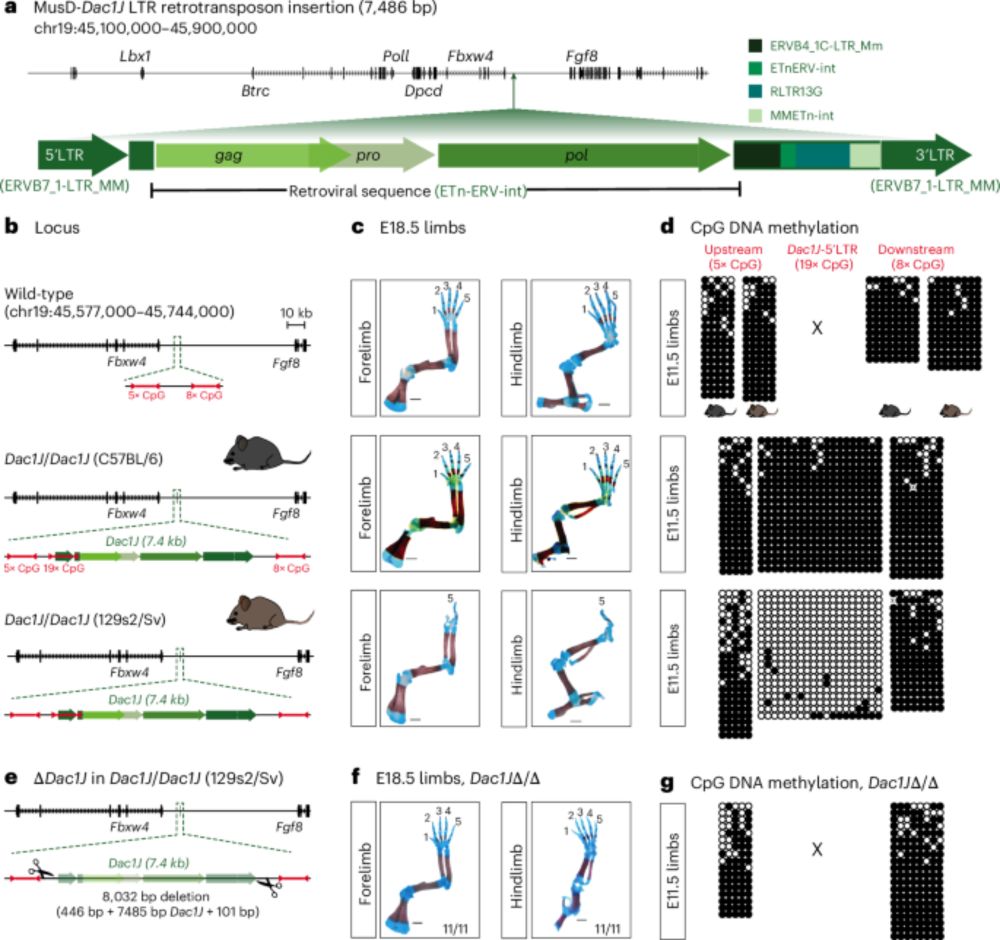
Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
(image from former PhD student Laura Cook)


(image from former PhD student Laura Cook)
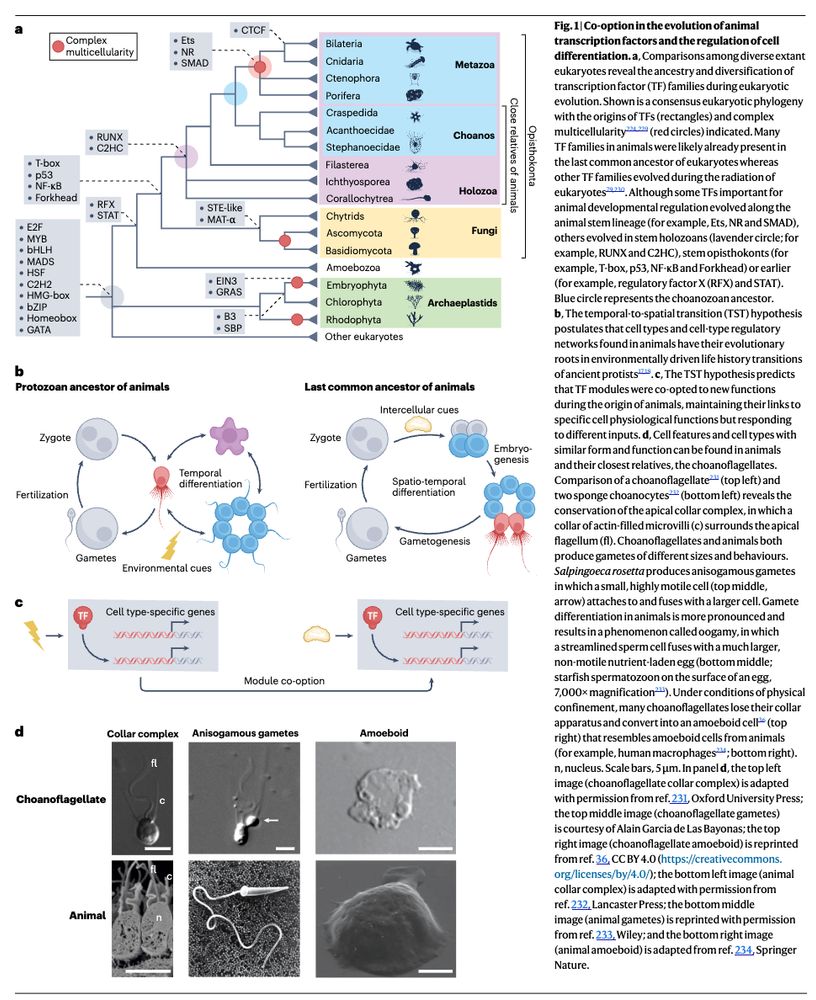
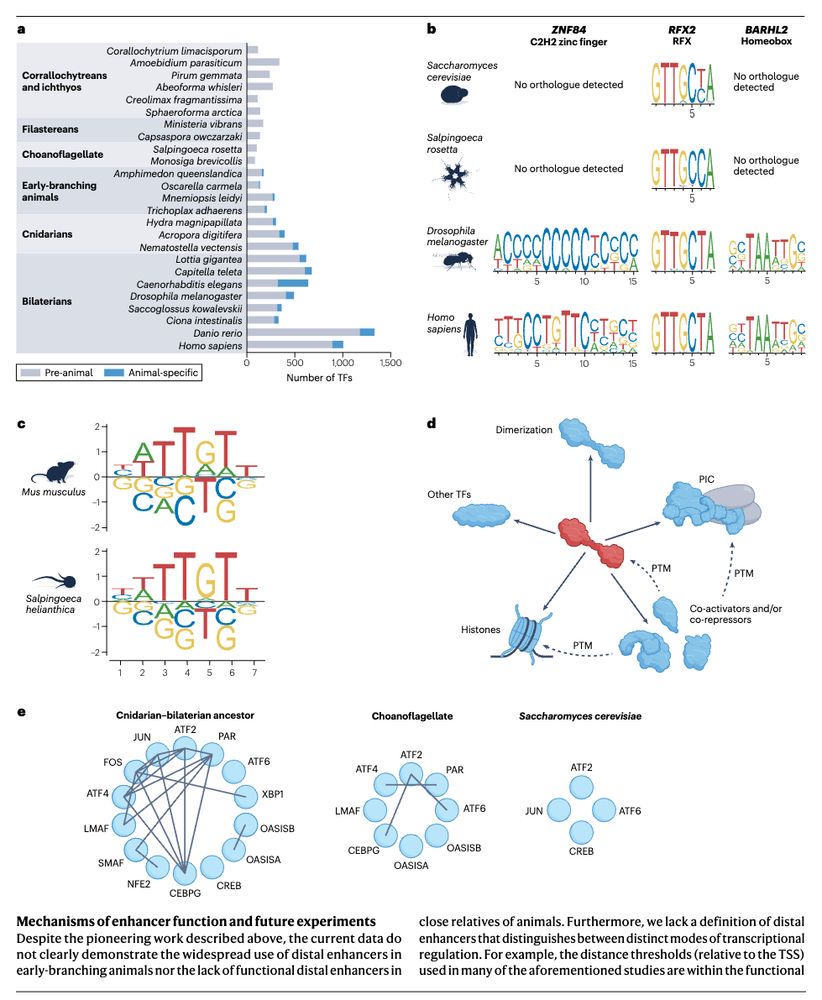
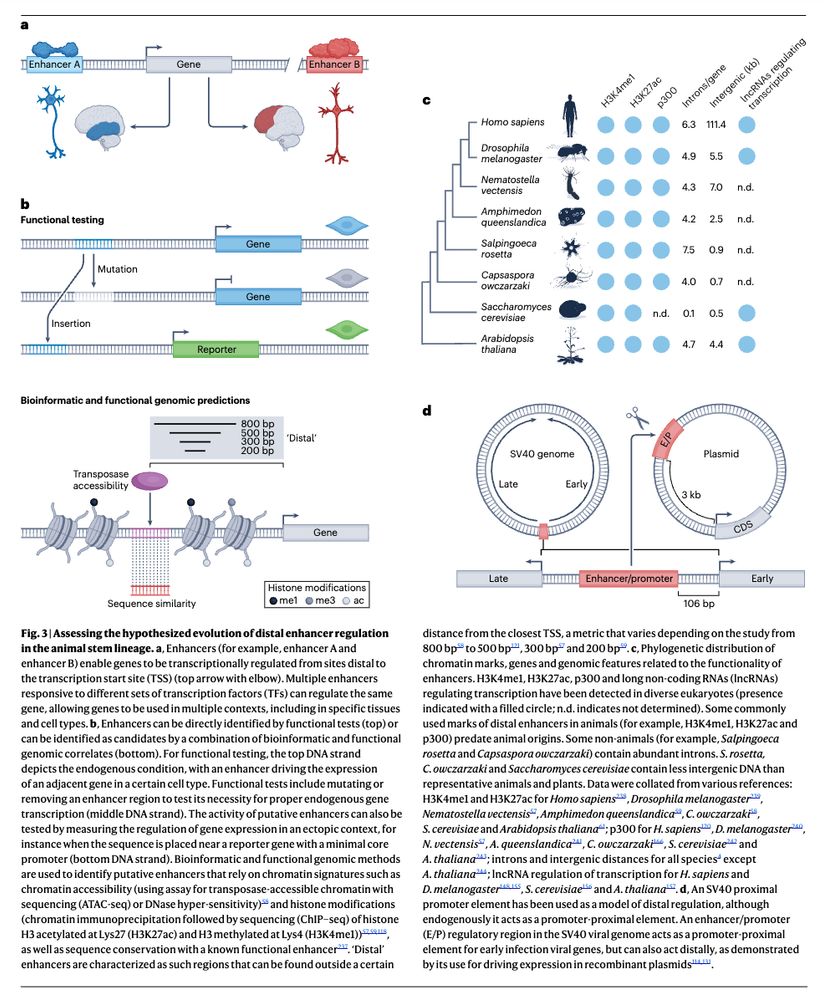




Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro

Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
You can register at: us06web.zoom.us/webinar/regi...
You can register at: us06web.zoom.us/webinar/regi...
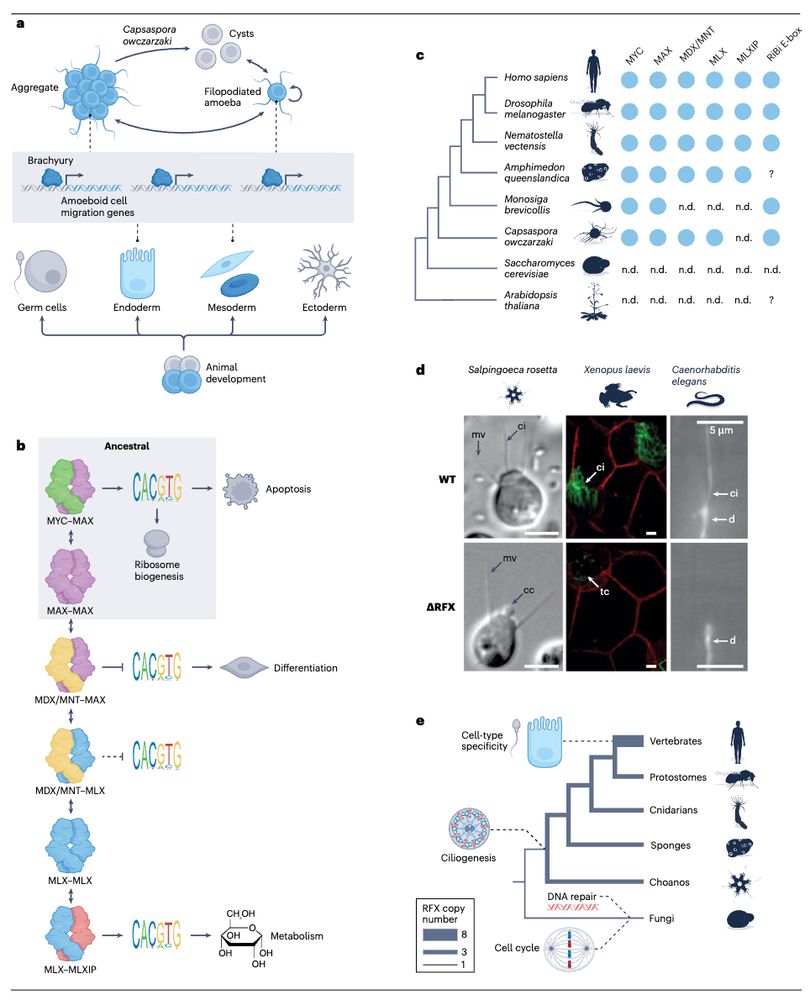
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
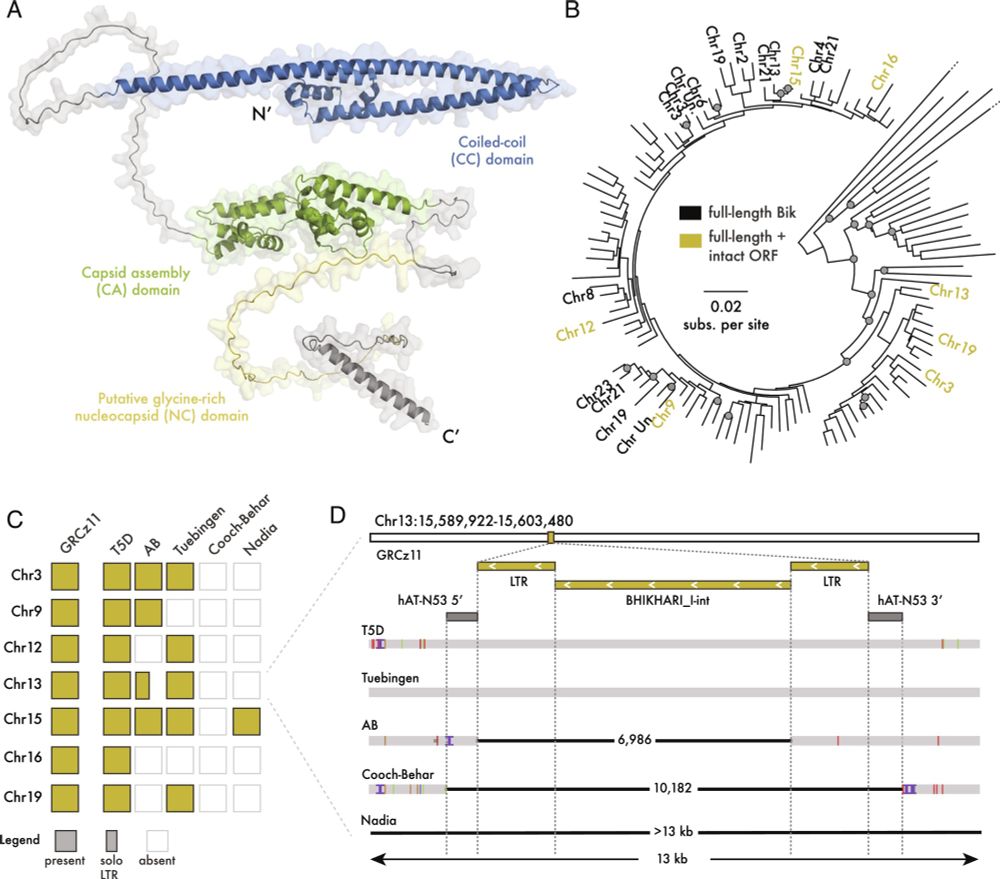
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
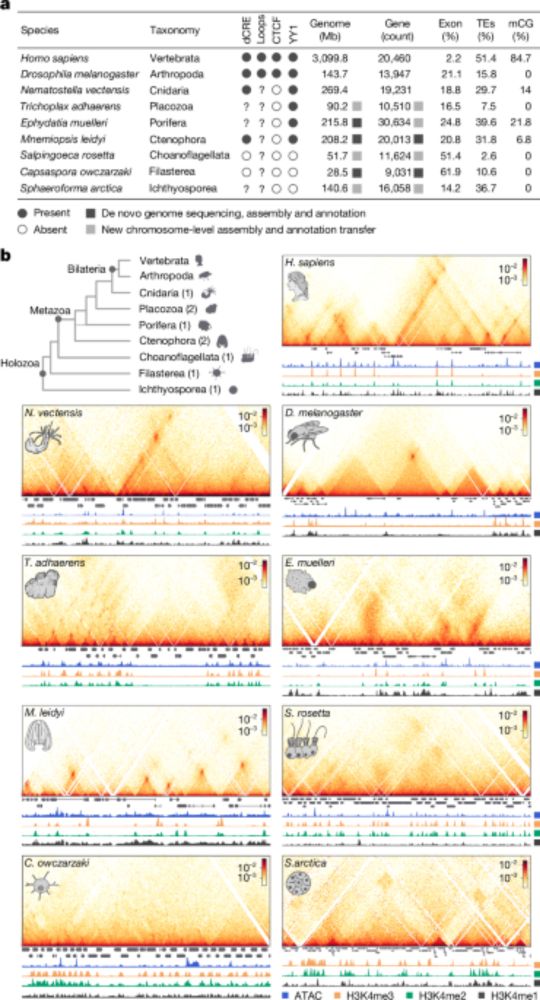
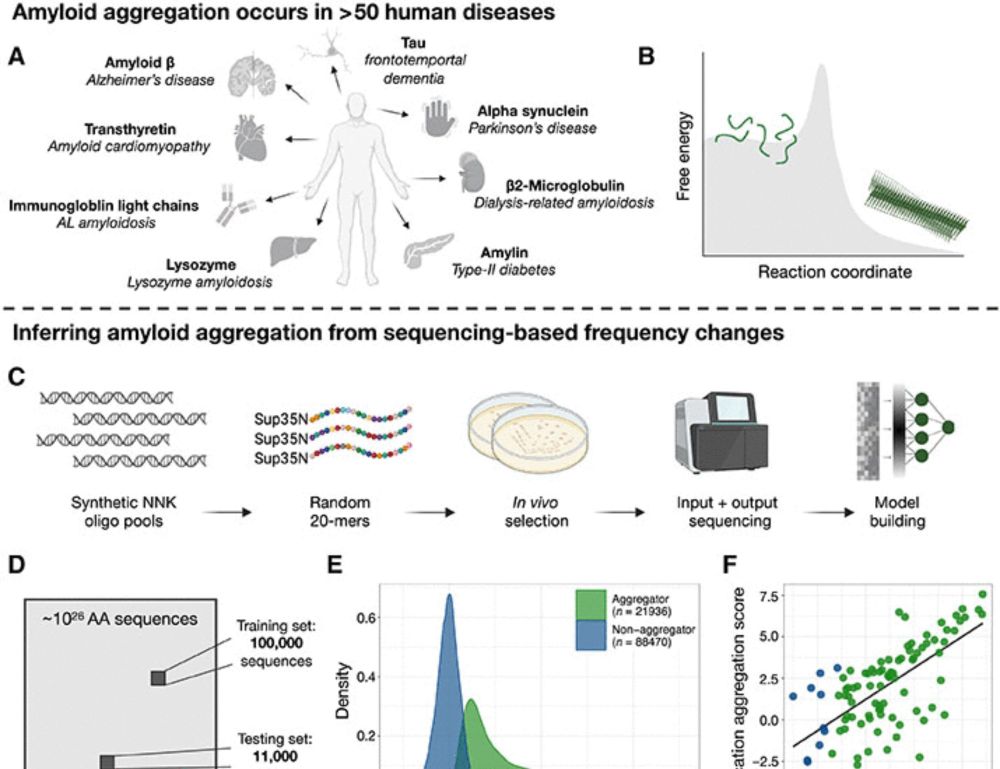

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...

Preprint: www.biorxiv.org/content/10.1...
GitHub: github.com/jmschrei/led...
🧬 "Enhancers Sequences and Gene Regulation"
🖥️ Les vidéos du colloque organisé par le Pr @denisduboule.bsky.social, titulaire de la chaire Évolution du développement et des génomes, sont disponibles.
👉 www.college-de-france.fr/fr/agenda/co...

4-5 PM - Bloedel Conservatory (Free Entry)
5 PM - BBQ at QE Park
Feel free to join for both parts of the event or just one.
Register now: ubc.ca1.qualtrics.com/jfe/form/SV_...
4-5 PM - Bloedel Conservatory (Free Entry)
5 PM - BBQ at QE Park
Feel free to join for both parts of the event or just one.
Register now: ubc.ca1.qualtrics.com/jfe/form/SV_...
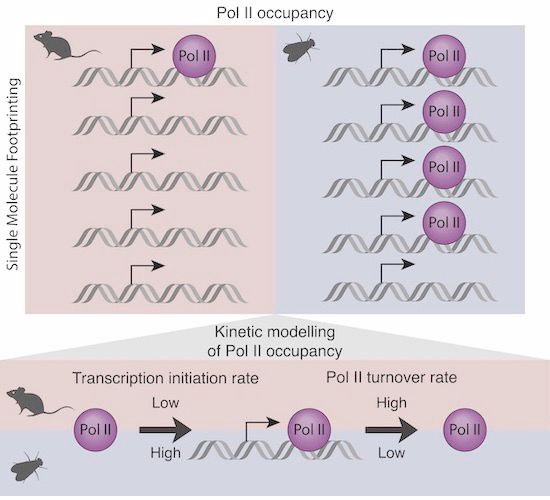
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

