

read the first PLUMB blab of many below, and follow along as Ariana builds an open comp.chem project from the ground up.
read the first PLUMB blab of many below, and follow along as Ariana builds an open comp.chem project from the ground up.

pubs.acs.org/doi/full/10....

pubs.acs.org/doi/full/10....

Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
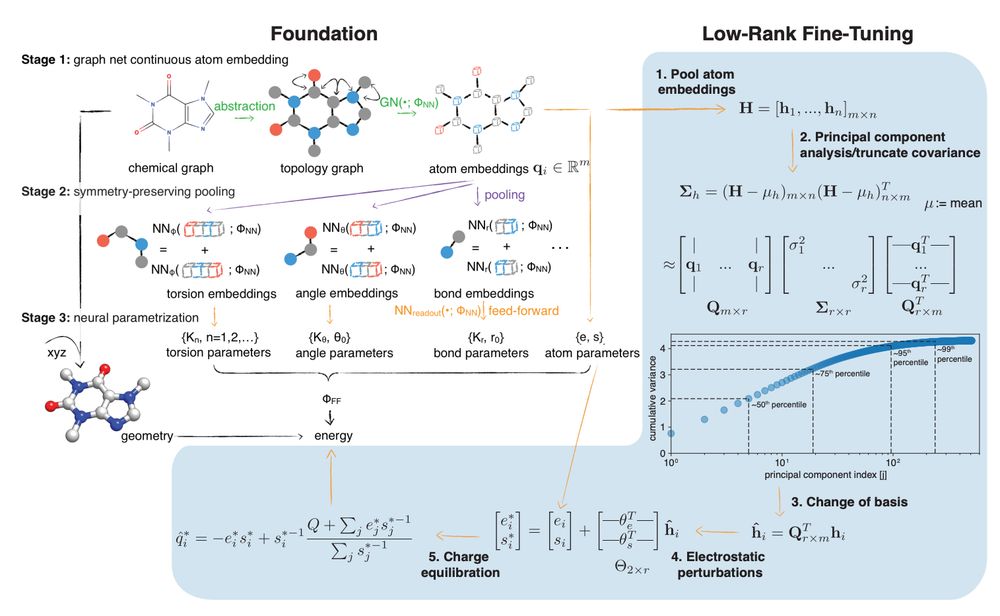
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
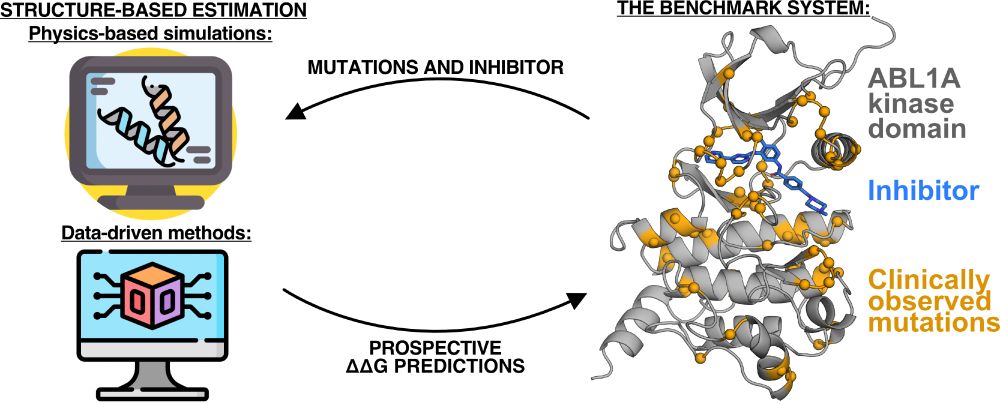
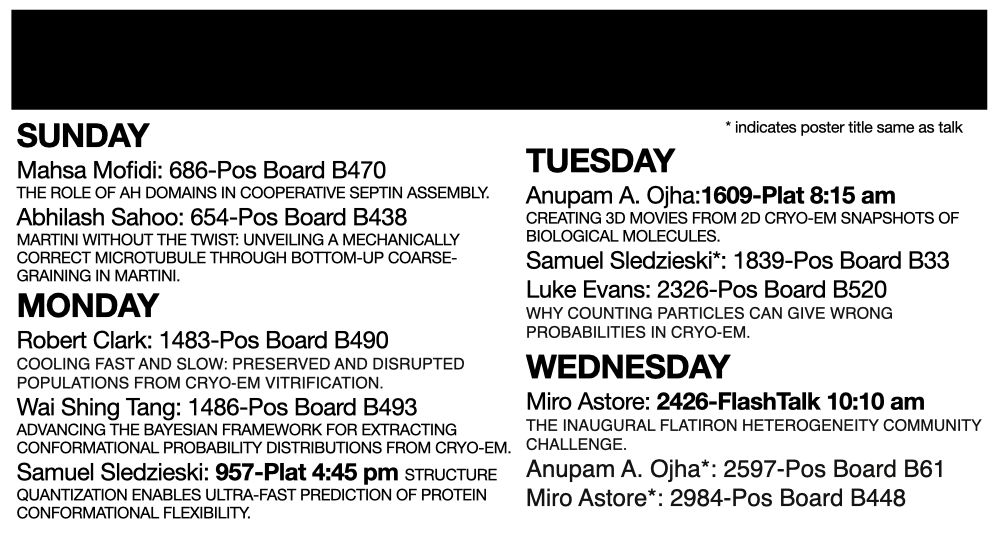
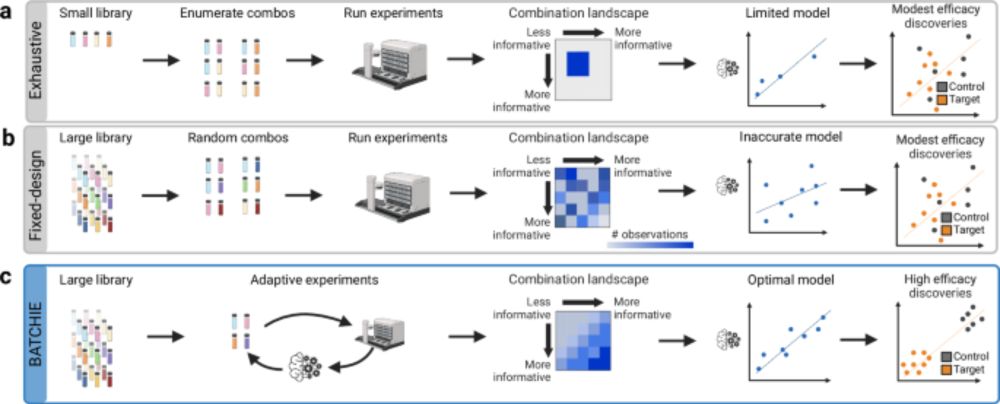
2024 was a productive year, and 2025 stands to be even more exciting!
Our thanks to everyone who helped, and some reflections: foldingathome.org/2025/01/09/h...
2024 was a productive year, and 2025 stands to be even more exciting!
Our thanks to everyone who helped, and some reflections: foldingathome.org/2025/01/09/h...

Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...
Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...

