
Previous: SWTX | OrbiMed | WeillCornellGS | BofAML | Penn | Andover
Employer-mandated disclaimer: choderalab.org/disclaimer
Apply at achira.ai

Apply at achira.ai
Accurate drug docking needs crystal struct. Template docking via mol. similarity helps w/ ltd. data.




Accurate drug docking needs crystal struct. Template docking via mol. similarity helps w/ ltd. data.
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT


Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
RSVP link dropping soon in all ACS COMP members' email inboxes :)


If you love college sports, you're going to have to stand up for colleges!!!!
If you love college sports, you're going to have to stand up for colleges!!!!
pubs.acs.org/doi/full/10....

pubs.acs.org/doi/full/10....

www.nytimes.com/2025/03/01/u...

www.nytimes.com/2025/03/01/u...
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
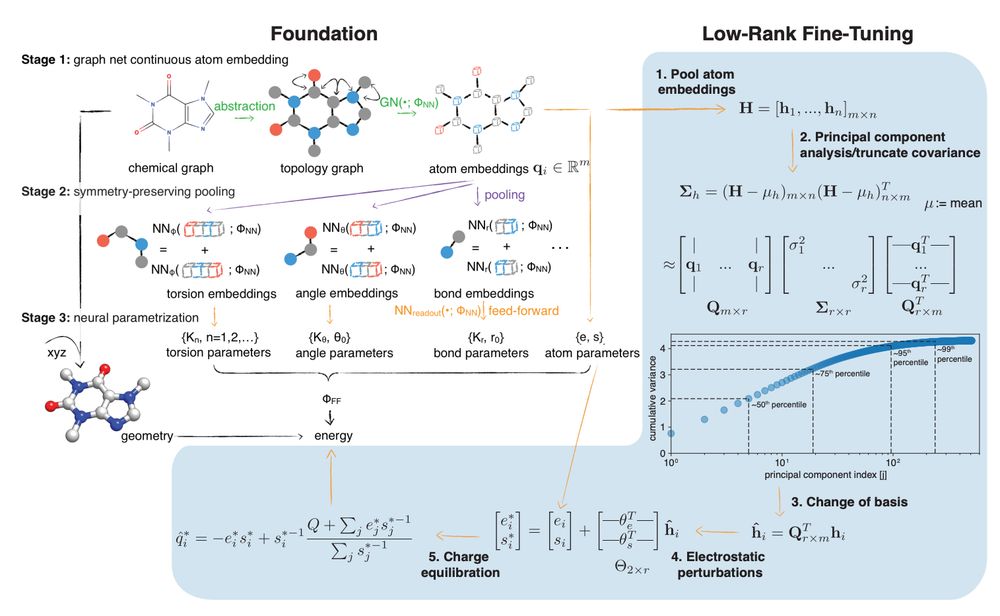
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.

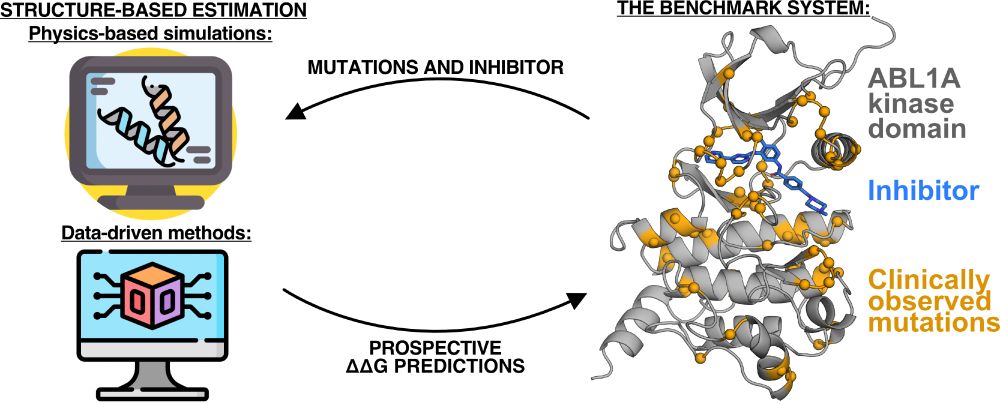
I want to provide some context on what that means and why it matters.
grants.nih.gov/grants/guide...
I want to provide some context on what that means and why it matters.
grants.nih.gov/grants/guide...
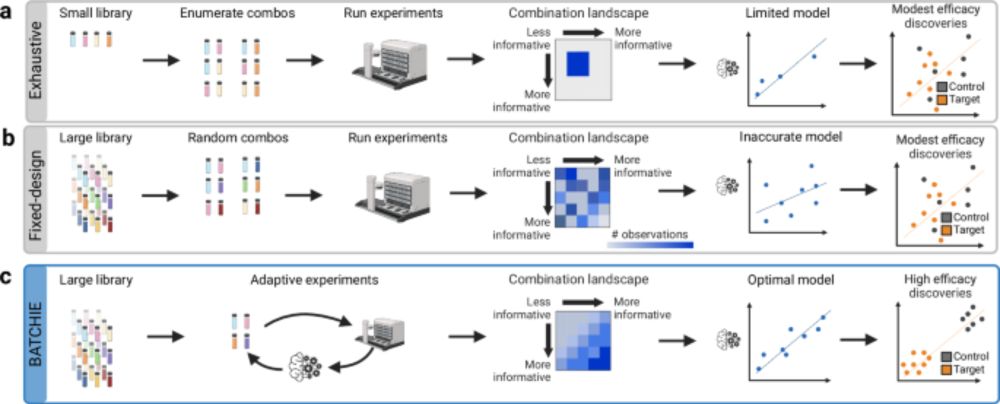
Ready to test your skills on new data? Hosted in partnership with @asapdiscovery.bsky.social and @omsf.io, we've prepared detailed notebooks showcasing how to format your data and submit your solutions. 🧑💻

Ready to test your skills on new data? Hosted in partnership with @asapdiscovery.bsky.social and @omsf.io, we've prepared detailed notebooks showcasing how to format your data and submit your solutions. 🧑💻
Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...
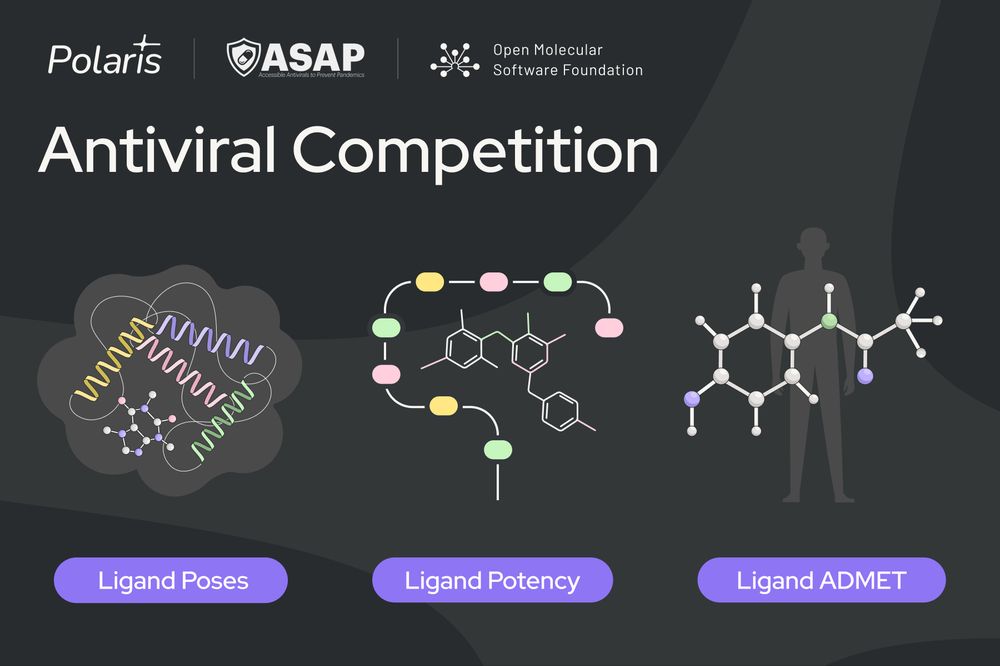
Test your skills across three sub-challenges revolving around SARS-CoV-2 and MERS-CoV Mpro🧵
Full details: polarishub.io/competitions
Blog: polarishub.io/blog/antivir...
www.importantcontext.news/p/judges-kee...

www.importantcontext.news/p/judges-kee...



