
computational biophysicist, glycoscientist, feminist, jewish, lover of sufjan stevens. she/they 🏳️🌈 ✡︎
📍 sf/nyc

currently a biophysicist-turned-ML-scientist @ iambic tx, developing & utilizing structure prediction algorithms in drug disco
prev: phd w/ @rommieamaro.bsky.social, postdoc w/ bernie brooks & @jchodera.bsky.social 🤓
RSVP link dropping soon in all ACS COMP members' email inboxes :)

It has been running continuously since 1991 and has provided massive key knowledge about diseases in women
Unreal
www.science.org/content/arti...

It has been running continuously since 1991 and has provided massive key knowledge about diseases in women
Unreal
www.science.org/content/arti...
This free virtual course will demonstrate the power and flexibility of python on May 1, 2025
pdb101.rcsb.org/news...

This free virtual course will demonstrate the power and flexibility of python on May 1, 2025
pdb101.rcsb.org/news...

According to a blog post on the company’s website, OpenAI raised $40 billion in a round that values the company at $300 billion post-money.
Read more here: tcrn.ch/4lbbzCf

cen.acs.org/acs-news/acs...

cen.acs.org/acs-news/acs...
Are you a comp chem trainee attending Spring ACS in San Diego? Would you like to expand your network with mentors in industry & academia at the COMP Mentor Lunch?
Fill out this form if you are interested in attending!
forms.gle/Sq79TJihHnFW...
Are you a comp chem trainee attending Spring ACS in San Diego? Would you like to expand your network with mentors in industry & academia at the COMP Mentor Lunch?
Fill out this form if you are interested in attending!
forms.gle/Sq79TJihHnFW...

Hannah Wayment-Steele
@ginaelnesr.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...




Hannah Wayment-Steele
@ginaelnesr.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
📣 NEW BIORXIV ALERT!! 🚨
Using WE MD, linguistic pathway clustering, dynamical network analyses, and HDXMS we reveal a hidden allosteric network within the SARS2 spike S1 domain and predict how the D614G mutation impacts this network!
www.biorxiv.org/content/10.1...

hope to see y'all there!!!
If you didn’t get the RSVP link in your email inbox, DM us with a screenshot of your COMP membership and we’ll shoot you the RSVP link 😁
#ACSSpring2025 #chemsky

hope to see y'all there!!!


Speak up. Stand up. Get out there and protect out democracy
Keep an eye on this space for updates, event information, and ways to get involved. We can't wait to see everyone #standupforscience2025 on March 7th, both in DC and locations nationwide!
#scienceforall #sciencenotsilence

Speak up. Stand up. Get out there and protect out democracy
pls share with folks you think may be interested, & feel free to reach out with questions
jobs.ashbyhq.com/iambic-thera...

pls share with folks you think may be interested, & feel free to reach out with questions
jobs.ashbyhq.com/iambic-thera...
research.arcadiascience.com/pub/result-p...
research.arcadiascience.com/pub/result-p...
if you're interested in the evaluation of conformational accuracy of structure prediction methods, take a look at our first stab at a systematic conformational benchmark in the NP3 technical report below! 🧵
www.iambic.ai/post/np3-tec...
if you're interested in the evaluation of conformational accuracy of structure prediction methods, take a look at our first stab at a systematic conformational benchmark in the NP3 technical report below! 🧵
www.iambic.ai/post/np3-tec...
Friends don't let friends use boxplot for binomial (bimodal) data. Is your box plot hiding something from you?
#DataVisualization
github.com/cxli233/Frie...
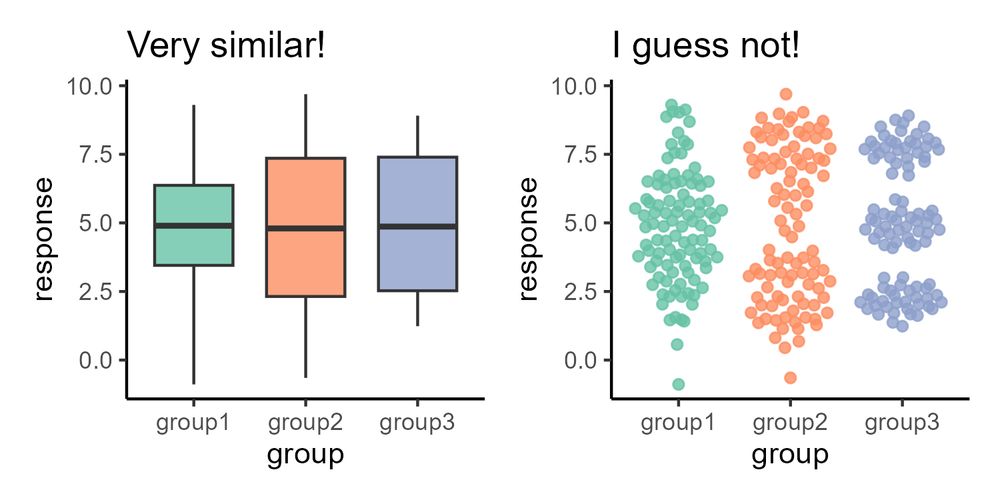
Friends don't let friends use boxplot for binomial (bimodal) data. Is your box plot hiding something from you?
#DataVisualization
github.com/cxli233/Frie...
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
www.thelancet.com/journals/lan...




